
Cicer arietinum (Chickpea) (Garbanzo)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; 50 kb inversion clade; NPAAA clade; Hologalegina;
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
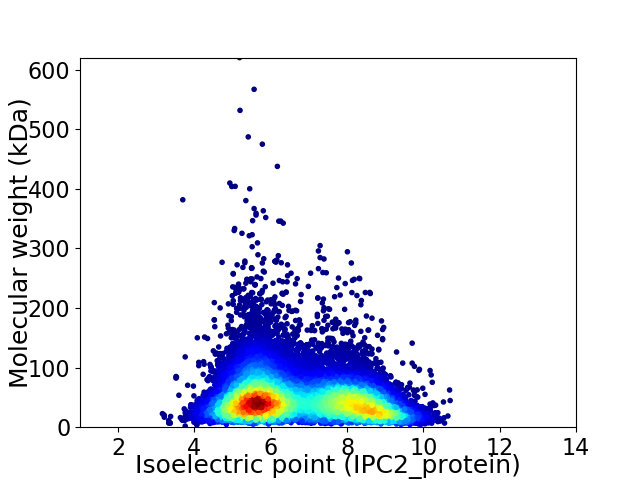
Virtual 2D-PAGE plot for 30622 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
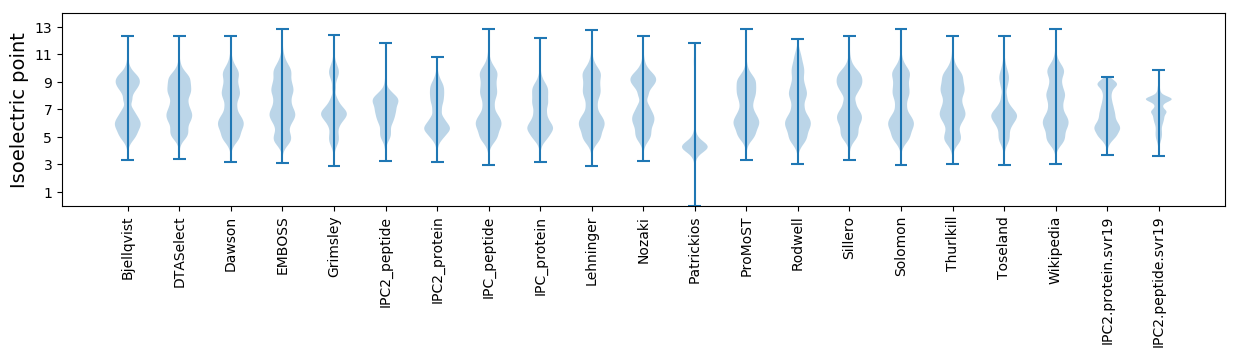
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q7Y697|A0A3Q7Y697_CICAR Isoform of A0A3Q7XQD1 B3 domain-containing protein LOC_Os12g40080-like isoform X2 OS=Cicer arietinum OX=3827 GN=LOC101504675 PE=4 SV=1
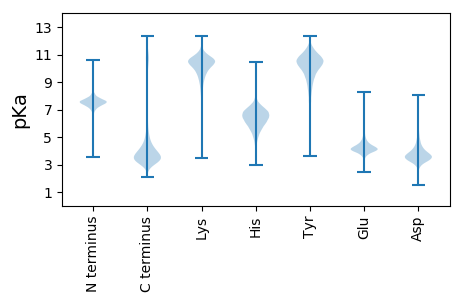
MM1 pKa = 7.68EE2 pKa = 5.67LPEE5 pKa = 4.86PPTGFVLDD13 pKa = 4.41DD14 pKa = 4.11NGTVTLSTTNRR25 pKa = 11.84LATIVDD31 pKa = 5.11PISNLPLEE39 pKa = 4.39CVIRR43 pKa = 11.84RR44 pKa = 11.84VFKK47 pKa = 11.0SSDD50 pKa = 3.23RR51 pKa = 11.84DD52 pKa = 3.34EE53 pKa = 5.72CMLLCPVDD61 pKa = 3.83TPVQILKK68 pKa = 8.43STRR71 pKa = 11.84DD72 pKa = 3.13GWSAISDD79 pKa = 3.84EE80 pKa = 4.52EE81 pKa = 4.51DD82 pKa = 3.64EE83 pKa = 4.76SVLPAAAYY91 pKa = 10.03ALAKK95 pKa = 10.06IHH97 pKa = 5.5MHH99 pKa = 6.11LVYY102 pKa = 10.55SGYY105 pKa = 10.69CYY107 pKa = 8.81TARR110 pKa = 11.84GGFCYY115 pKa = 9.84TDD117 pKa = 3.76KK118 pKa = 11.16DD119 pKa = 3.41ILDD122 pKa = 3.74FHH124 pKa = 6.76TDD126 pKa = 3.2DD127 pKa = 4.01GKK129 pKa = 11.28EE130 pKa = 3.97VDD132 pKa = 4.09GLPSEE137 pKa = 4.5GVEE140 pKa = 3.41ITYY143 pKa = 9.61FYY145 pKa = 11.06LKK147 pKa = 8.62NTRR150 pKa = 11.84YY151 pKa = 10.13LIYY154 pKa = 10.01TPPEE158 pKa = 3.7PVQFVVVKK166 pKa = 10.46DD167 pKa = 3.9EE168 pKa = 4.29NGMFQMADD176 pKa = 3.92DD177 pKa = 5.39DD178 pKa = 5.33LLDD181 pKa = 4.61DD182 pKa = 4.68PAVIDD187 pKa = 4.91SIDD190 pKa = 3.43EE191 pKa = 4.07EE192 pKa = 5.09TEE194 pKa = 3.85FNALVEE200 pKa = 4.23EE201 pKa = 4.25EE202 pKa = 4.26AAVIEE207 pKa = 4.18ALMDD211 pKa = 4.08DD212 pKa = 4.76EE213 pKa = 5.02EE214 pKa = 6.48DD215 pKa = 3.71SDD217 pKa = 4.79EE218 pKa = 4.22EE219 pKa = 4.72DD220 pKa = 4.34YY221 pKa = 11.5YY222 pKa = 12.01
MM1 pKa = 7.68EE2 pKa = 5.67LPEE5 pKa = 4.86PPTGFVLDD13 pKa = 4.41DD14 pKa = 4.11NGTVTLSTTNRR25 pKa = 11.84LATIVDD31 pKa = 5.11PISNLPLEE39 pKa = 4.39CVIRR43 pKa = 11.84RR44 pKa = 11.84VFKK47 pKa = 11.0SSDD50 pKa = 3.23RR51 pKa = 11.84DD52 pKa = 3.34EE53 pKa = 5.72CMLLCPVDD61 pKa = 3.83TPVQILKK68 pKa = 8.43STRR71 pKa = 11.84DD72 pKa = 3.13GWSAISDD79 pKa = 3.84EE80 pKa = 4.52EE81 pKa = 4.51DD82 pKa = 3.64EE83 pKa = 4.76SVLPAAAYY91 pKa = 10.03ALAKK95 pKa = 10.06IHH97 pKa = 5.5MHH99 pKa = 6.11LVYY102 pKa = 10.55SGYY105 pKa = 10.69CYY107 pKa = 8.81TARR110 pKa = 11.84GGFCYY115 pKa = 9.84TDD117 pKa = 3.76KK118 pKa = 11.16DD119 pKa = 3.41ILDD122 pKa = 3.74FHH124 pKa = 6.76TDD126 pKa = 3.2DD127 pKa = 4.01GKK129 pKa = 11.28EE130 pKa = 3.97VDD132 pKa = 4.09GLPSEE137 pKa = 4.5GVEE140 pKa = 3.41ITYY143 pKa = 9.61FYY145 pKa = 11.06LKK147 pKa = 8.62NTRR150 pKa = 11.84YY151 pKa = 10.13LIYY154 pKa = 10.01TPPEE158 pKa = 3.7PVQFVVVKK166 pKa = 10.46DD167 pKa = 3.9EE168 pKa = 4.29NGMFQMADD176 pKa = 3.92DD177 pKa = 5.39DD178 pKa = 5.33LLDD181 pKa = 4.61DD182 pKa = 4.68PAVIDD187 pKa = 4.91SIDD190 pKa = 3.43EE191 pKa = 4.07EE192 pKa = 5.09TEE194 pKa = 3.85FNALVEE200 pKa = 4.23EE201 pKa = 4.25EE202 pKa = 4.26AAVIEE207 pKa = 4.18ALMDD211 pKa = 4.08DD212 pKa = 4.76EE213 pKa = 5.02EE214 pKa = 6.48DD215 pKa = 3.71SDD217 pKa = 4.79EE218 pKa = 4.22EE219 pKa = 4.72DD220 pKa = 4.34YY221 pKa = 11.5YY222 pKa = 12.01
Molecular weight: 24.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S2YD42|A0A1S2YD42_CICAR CASP-like protein OS=Cicer arietinum OX=3827 GN=LOC101515680 PE=3 SV=1
MM1 pKa = 7.18FGSVRR6 pKa = 11.84IVSNIAACIRR16 pKa = 11.84QNLSLHH22 pKa = 5.88GVQVRR27 pKa = 11.84NINIGGGLGGEE38 pKa = 4.27IPDD41 pKa = 4.25SKK43 pKa = 10.55RR44 pKa = 11.84LQYY47 pKa = 11.3ALQHH51 pKa = 5.12LHH53 pKa = 5.97GVGRR57 pKa = 11.84SKK59 pKa = 10.81AHH61 pKa = 6.88HH62 pKa = 6.25IVCEE66 pKa = 4.03LGVEE70 pKa = 4.11NKK72 pKa = 9.69YY73 pKa = 11.19VKK75 pKa = 10.43DD76 pKa = 3.54LSKK79 pKa = 11.16RR80 pKa = 11.84EE81 pKa = 4.04LYY83 pKa = 10.4SLRR86 pKa = 11.84EE87 pKa = 4.13LLSKK91 pKa = 10.61YY92 pKa = 10.85LIGNDD97 pKa = 3.18LKK99 pKa = 11.29KK100 pKa = 10.69LVEE103 pKa = 4.23RR104 pKa = 11.84DD105 pKa = 3.05VGRR108 pKa = 11.84LVGIQCYY115 pKa = 9.89RR116 pKa = 11.84GIRR119 pKa = 11.84HH120 pKa = 6.11VDD122 pKa = 3.25GLPCRR127 pKa = 11.84GQRR130 pKa = 11.84THH132 pKa = 6.03TNSRR136 pKa = 11.84TRR138 pKa = 11.84RR139 pKa = 11.84TMRR142 pKa = 11.84TFGGSRR148 pKa = 3.16
MM1 pKa = 7.18FGSVRR6 pKa = 11.84IVSNIAACIRR16 pKa = 11.84QNLSLHH22 pKa = 5.88GVQVRR27 pKa = 11.84NINIGGGLGGEE38 pKa = 4.27IPDD41 pKa = 4.25SKK43 pKa = 10.55RR44 pKa = 11.84LQYY47 pKa = 11.3ALQHH51 pKa = 5.12LHH53 pKa = 5.97GVGRR57 pKa = 11.84SKK59 pKa = 10.81AHH61 pKa = 6.88HH62 pKa = 6.25IVCEE66 pKa = 4.03LGVEE70 pKa = 4.11NKK72 pKa = 9.69YY73 pKa = 11.19VKK75 pKa = 10.43DD76 pKa = 3.54LSKK79 pKa = 11.16RR80 pKa = 11.84EE81 pKa = 4.04LYY83 pKa = 10.4SLRR86 pKa = 11.84EE87 pKa = 4.13LLSKK91 pKa = 10.61YY92 pKa = 10.85LIGNDD97 pKa = 3.18LKK99 pKa = 11.29KK100 pKa = 10.69LVEE103 pKa = 4.23RR104 pKa = 11.84DD105 pKa = 3.05VGRR108 pKa = 11.84LVGIQCYY115 pKa = 9.89RR116 pKa = 11.84GIRR119 pKa = 11.84HH120 pKa = 6.11VDD122 pKa = 3.25GLPCRR127 pKa = 11.84GQRR130 pKa = 11.84THH132 pKa = 6.03TNSRR136 pKa = 11.84TRR138 pKa = 11.84RR139 pKa = 11.84TMRR142 pKa = 11.84TFGGSRR148 pKa = 3.16
Molecular weight: 16.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
14024822 |
20 |
5450 |
458.0 |
51.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.099 ± 0.012 | 1.872 ± 0.007 |
5.361 ± 0.009 | 6.413 ± 0.017 |
4.311 ± 0.009 | 6.181 ± 0.017 |
2.522 ± 0.006 | 5.685 ± 0.01 |
6.403 ± 0.013 | 9.588 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.005 | 5.003 ± 0.01 |
4.726 ± 0.016 | 3.763 ± 0.01 |
4.942 ± 0.011 | 9.084 ± 0.017 |
5.029 ± 0.008 | 6.452 ± 0.009 |
1.244 ± 0.004 | 2.897 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
