
Malonomonas rubra DSM 5091
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Desulfuromonadaceae; Malonomonas; Malonomonas rubra
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
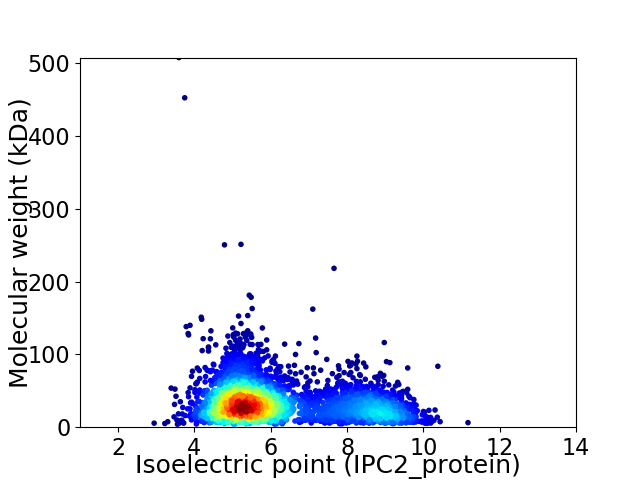
Virtual 2D-PAGE plot for 3654 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6IQ36|A0A1M6IQ36_MALRU Multidrug efflux pump subunit AcrB OS=Malonomonas rubra DSM 5091 OX=1122189 GN=SAMN02745165_02201 PE=4 SV=1
MM1 pKa = 7.95KK2 pKa = 10.19YY3 pKa = 10.29NPLIIIFGMLLLLPQVGYY21 pKa = 10.01SAQDD25 pKa = 3.28SYY27 pKa = 11.76IGDD30 pKa = 3.53TAIYY34 pKa = 9.63GGASVTLQPNVLIVFDD50 pKa = 4.55TSGSMDD56 pKa = 4.13DD57 pKa = 4.44EE58 pKa = 6.12IDD60 pKa = 3.9VQVCQPDD67 pKa = 3.46VDD69 pKa = 4.01SDD71 pKa = 4.19GVIDD75 pKa = 5.33EE76 pKa = 5.05SDD78 pKa = 3.55NCPTVANATQTDD90 pKa = 3.64TDD92 pKa = 3.52NDD94 pKa = 5.21GIGDD98 pKa = 4.11LCDD101 pKa = 4.97DD102 pKa = 3.86NTTFPDD108 pKa = 3.48TDD110 pKa = 3.78GDD112 pKa = 4.38GVTDD116 pKa = 3.81DD117 pKa = 4.25TDD119 pKa = 3.55NCKK122 pKa = 8.94VTSNADD128 pKa = 3.37QADD131 pKa = 3.67SDD133 pKa = 4.2GDD135 pKa = 4.34GIGDD139 pKa = 3.59ACEE142 pKa = 4.18VGSGGYY148 pKa = 10.13NKK150 pKa = 9.63DD151 pKa = 2.93TDD153 pKa = 4.07YY154 pKa = 11.0TAMTSSEE161 pKa = 4.09FCGDD165 pKa = 2.84GGGRR169 pKa = 11.84RR170 pKa = 11.84DD171 pKa = 3.9PPWAAEE177 pKa = 3.63ACQRR181 pKa = 11.84NKK183 pKa = 10.12VYY185 pKa = 10.57RR186 pKa = 11.84CRR188 pKa = 11.84DD189 pKa = 3.76DD190 pKa = 4.96EE191 pKa = 4.04WDD193 pKa = 3.47QNGFCSQWDD202 pKa = 3.46RR203 pKa = 11.84VEE205 pKa = 4.62NIDD208 pKa = 5.63YY209 pKa = 11.24SDD211 pKa = 3.85IDD213 pKa = 4.65CNDD216 pKa = 3.16QEE218 pKa = 5.2DD219 pKa = 4.12ALSEE223 pKa = 4.01QGTLVDD229 pKa = 5.05DD230 pKa = 4.01VRR232 pKa = 11.84FDD234 pKa = 4.08EE235 pKa = 6.24DD236 pKa = 4.47GNCDD240 pKa = 3.08GSRR243 pKa = 11.84SSRR246 pKa = 11.84YY247 pKa = 8.66YY248 pKa = 10.32ATGNWVDD255 pKa = 3.47WYY257 pKa = 11.3SLVTIGDD264 pKa = 3.7AGGGASGSGDD274 pKa = 3.79DD275 pKa = 4.27YY276 pKa = 11.82NSGSTQICSTTQEE289 pKa = 4.13RR290 pKa = 11.84KK291 pKa = 9.9NDD293 pKa = 3.26VAEE296 pKa = 4.22NVVSDD301 pKa = 5.01LIEE304 pKa = 4.13STGGVNFGIMRR315 pKa = 11.84FNSSSGGTFMTVSVDD330 pKa = 3.21GQNYY334 pKa = 6.38EE335 pKa = 4.25TTIKK339 pKa = 10.99NMDD342 pKa = 4.45EE343 pKa = 3.8IHH345 pKa = 6.66TGTTTNRR352 pKa = 11.84EE353 pKa = 4.1ALVDD357 pKa = 3.47IASNMPASGYY367 pKa = 8.6TPLGEE372 pKa = 4.29TLYY375 pKa = 11.0EE376 pKa = 3.85AMRR379 pKa = 11.84YY380 pKa = 8.82FSGGNKK386 pKa = 9.18YY387 pKa = 9.91FAGSGSYY394 pKa = 9.29TSPITASCQPNYY406 pKa = 9.85IILMTDD412 pKa = 2.98GMATSDD418 pKa = 3.5SSVPSLSCGGDD429 pKa = 3.54DD430 pKa = 5.91NVDD433 pKa = 3.43CDD435 pKa = 4.04GDD437 pKa = 3.98GRR439 pKa = 11.84SDD441 pKa = 5.31DD442 pKa = 4.75GDD444 pKa = 4.51DD445 pKa = 5.3DD446 pKa = 5.5SLDD449 pKa = 3.63DD450 pKa = 4.31VAWYY454 pKa = 10.01LYY456 pKa = 9.37NTDD459 pKa = 4.52LSDD462 pKa = 4.65FNGTQNVKK470 pKa = 9.62TYY472 pKa = 10.28TIGFGLGGGDD482 pKa = 3.62SDD484 pKa = 6.07AVDD487 pKa = 4.8LLQDD491 pKa = 3.63TADD494 pKa = 3.77NGQGANAGQGKK505 pKa = 10.03SYY507 pKa = 10.38LASSYY512 pKa = 11.2QEE514 pKa = 3.87LTSALSAIIGEE525 pKa = 4.4VLDD528 pKa = 3.68QSSAFVAPVVPTSPEE543 pKa = 3.6NKK545 pKa = 8.99VYY547 pKa = 10.37SGQRR551 pKa = 11.84IYY553 pKa = 11.24LGFFKK558 pKa = 10.62PQITGDD564 pKa = 2.86WYY566 pKa = 11.33GNLKK570 pKa = 10.55KK571 pKa = 10.77FGLDD575 pKa = 3.29NNGEE579 pKa = 4.23VLDD582 pKa = 4.31KK583 pKa = 11.24NGNEE587 pKa = 3.85ATDD590 pKa = 3.65ANGAFLPEE598 pKa = 4.35SVSYY602 pKa = 10.08WGTATDD608 pKa = 3.43AGNVEE613 pKa = 4.27EE614 pKa = 5.05GGIGGILNNRR624 pKa = 11.84VLSTRR629 pKa = 11.84NIYY632 pKa = 9.94TYY634 pKa = 10.59TGTTAALTDD643 pKa = 3.65SSNALTTSNTALTFADD659 pKa = 4.13FDD661 pKa = 3.94VASDD665 pKa = 3.58VRR667 pKa = 11.84KK668 pKa = 10.61DD669 pKa = 3.47EE670 pKa = 4.69IINYY674 pKa = 8.93VYY676 pKa = 10.7GYY678 pKa = 10.34DD679 pKa = 3.34VYY681 pKa = 11.25GINPSVGRR689 pKa = 11.84EE690 pKa = 3.7WVMGDD695 pKa = 3.06ILHH698 pKa = 6.65SKK700 pKa = 9.78PAIQTYY706 pKa = 10.4NSYY709 pKa = 11.15ALSSEE714 pKa = 3.97ADD716 pKa = 3.44PNVNKK721 pKa = 9.58TVIYY725 pKa = 10.43VGSNDD730 pKa = 3.73GQLHH734 pKa = 6.35AFSDD738 pKa = 4.39ANGSEE743 pKa = 4.25LWSFVPPSVLPNLQNLGNNAIHH765 pKa = 7.02EE766 pKa = 4.43YY767 pKa = 11.07FMDD770 pKa = 5.09GSPALYY776 pKa = 10.65VFDD779 pKa = 4.62YY780 pKa = 11.25DD781 pKa = 3.57GDD783 pKa = 4.3GNIGPGPEE791 pKa = 4.64SGDD794 pKa = 3.46TDD796 pKa = 3.68PTGVTDD802 pKa = 5.13AGANDD807 pKa = 3.91KK808 pKa = 11.22VIMVVSMRR816 pKa = 11.84RR817 pKa = 11.84GGGIDD822 pKa = 3.4TLDD825 pKa = 3.61AATSRR830 pKa = 11.84GSYY833 pKa = 9.5YY834 pKa = 11.06ALDD837 pKa = 3.66VTNPLSPQFLWEE849 pKa = 4.12IDD851 pKa = 3.48STTTGFSEE859 pKa = 5.72LGEE862 pKa = 4.3TWSDD866 pKa = 3.11AVIGKK871 pKa = 8.97IRR873 pKa = 11.84LNGSDD878 pKa = 4.54KK879 pKa = 10.5IVAFIGAGYY888 pKa = 10.52DD889 pKa = 3.37NNEE892 pKa = 4.29DD893 pKa = 3.46LRR895 pKa = 11.84FGNTQLFPDD904 pKa = 3.93NTTTATSTILPTADD918 pKa = 3.67ANDD921 pKa = 3.74VTSSGSSAQVNPKK934 pKa = 10.01GRR936 pKa = 11.84GIYY939 pKa = 9.97VIEE942 pKa = 4.58LATVSYY948 pKa = 8.13PTISPFLPTVAVNTSPQKK966 pKa = 9.68LWEE969 pKa = 4.1YY970 pKa = 11.25VYY972 pKa = 11.13DD973 pKa = 4.61ADD975 pKa = 6.28RR976 pKa = 11.84EE977 pKa = 4.17DD978 pKa = 4.49SNLDD982 pKa = 3.44AYY984 pKa = 11.0DD985 pKa = 4.0PGNNPKK991 pKa = 10.12YY992 pKa = 10.9SFVATIATLDD1002 pKa = 3.7SDD1004 pKa = 3.69FDD1006 pKa = 4.39GYY1008 pKa = 10.54IDD1010 pKa = 4.01RR1011 pKa = 11.84LYY1013 pKa = 11.31SGDD1016 pKa = 3.19TGGNMWRR1023 pKa = 11.84FDD1025 pKa = 3.66IGDD1028 pKa = 3.83KK1029 pKa = 10.44TSTSAWDD1036 pKa = 3.26ATKK1039 pKa = 10.33IFSANPSDD1047 pKa = 3.98VANSDD1052 pKa = 3.68EE1053 pKa = 4.96DD1054 pKa = 3.88PATNGRR1060 pKa = 11.84KK1061 pKa = 8.97IFYY1064 pKa = 9.69RR1065 pKa = 11.84PSIVQEE1071 pKa = 3.75PGYY1074 pKa = 10.27VGVYY1078 pKa = 10.29FGTGDD1083 pKa = 3.7RR1084 pKa = 11.84AHH1086 pKa = 7.5PLNKK1090 pKa = 10.09AVIDD1094 pKa = 3.82RR1095 pKa = 11.84LYY1097 pKa = 11.19AVYY1100 pKa = 10.54DD1101 pKa = 3.6RR1102 pKa = 11.84GNITSARR1109 pKa = 11.84TEE1111 pKa = 3.79EE1112 pKa = 4.18HH1113 pKa = 6.13LVNVTEE1119 pKa = 5.89DD1120 pKa = 3.73NLQSASPGIGVVNSTLLALSSTSNYY1145 pKa = 9.75GWFIKK1150 pKa = 10.59LDD1152 pKa = 3.84LLDD1155 pKa = 4.14GEE1157 pKa = 5.2KK1158 pKa = 10.72VLSNPIVFNKK1168 pKa = 8.58VAYY1171 pKa = 7.71YY1172 pKa = 7.36TTFTANVPSDD1182 pKa = 3.99DD1183 pKa = 3.98PCEE1186 pKa = 3.85QGQLGIGRR1194 pKa = 11.84LYY1196 pKa = 11.01AVDD1199 pKa = 4.01YY1200 pKa = 8.34KK1201 pKa = 10.91TGEE1204 pKa = 3.87AVFNFDD1210 pKa = 4.13LTNDD1214 pKa = 3.45VADD1217 pKa = 3.89EE1218 pKa = 4.15DD1219 pKa = 4.98SYY1221 pKa = 12.26AEE1223 pKa = 4.13NEE1225 pKa = 4.12NEE1227 pKa = 3.82RR1228 pKa = 11.84AIAGGGKK1235 pKa = 9.14ILRR1238 pKa = 11.84RR1239 pKa = 11.84SDD1241 pKa = 3.2RR1242 pKa = 11.84VKK1244 pKa = 10.21TIGSGIPSGAVVVVRR1259 pKa = 11.84EE1260 pKa = 4.78DD1261 pKa = 3.6GTSSALIGCGGGLCGGEE1278 pKa = 4.14TKK1280 pKa = 10.81GGGTAVPIYY1289 pKa = 10.74WMMEE1293 pKa = 3.59
MM1 pKa = 7.95KK2 pKa = 10.19YY3 pKa = 10.29NPLIIIFGMLLLLPQVGYY21 pKa = 10.01SAQDD25 pKa = 3.28SYY27 pKa = 11.76IGDD30 pKa = 3.53TAIYY34 pKa = 9.63GGASVTLQPNVLIVFDD50 pKa = 4.55TSGSMDD56 pKa = 4.13DD57 pKa = 4.44EE58 pKa = 6.12IDD60 pKa = 3.9VQVCQPDD67 pKa = 3.46VDD69 pKa = 4.01SDD71 pKa = 4.19GVIDD75 pKa = 5.33EE76 pKa = 5.05SDD78 pKa = 3.55NCPTVANATQTDD90 pKa = 3.64TDD92 pKa = 3.52NDD94 pKa = 5.21GIGDD98 pKa = 4.11LCDD101 pKa = 4.97DD102 pKa = 3.86NTTFPDD108 pKa = 3.48TDD110 pKa = 3.78GDD112 pKa = 4.38GVTDD116 pKa = 3.81DD117 pKa = 4.25TDD119 pKa = 3.55NCKK122 pKa = 8.94VTSNADD128 pKa = 3.37QADD131 pKa = 3.67SDD133 pKa = 4.2GDD135 pKa = 4.34GIGDD139 pKa = 3.59ACEE142 pKa = 4.18VGSGGYY148 pKa = 10.13NKK150 pKa = 9.63DD151 pKa = 2.93TDD153 pKa = 4.07YY154 pKa = 11.0TAMTSSEE161 pKa = 4.09FCGDD165 pKa = 2.84GGGRR169 pKa = 11.84RR170 pKa = 11.84DD171 pKa = 3.9PPWAAEE177 pKa = 3.63ACQRR181 pKa = 11.84NKK183 pKa = 10.12VYY185 pKa = 10.57RR186 pKa = 11.84CRR188 pKa = 11.84DD189 pKa = 3.76DD190 pKa = 4.96EE191 pKa = 4.04WDD193 pKa = 3.47QNGFCSQWDD202 pKa = 3.46RR203 pKa = 11.84VEE205 pKa = 4.62NIDD208 pKa = 5.63YY209 pKa = 11.24SDD211 pKa = 3.85IDD213 pKa = 4.65CNDD216 pKa = 3.16QEE218 pKa = 5.2DD219 pKa = 4.12ALSEE223 pKa = 4.01QGTLVDD229 pKa = 5.05DD230 pKa = 4.01VRR232 pKa = 11.84FDD234 pKa = 4.08EE235 pKa = 6.24DD236 pKa = 4.47GNCDD240 pKa = 3.08GSRR243 pKa = 11.84SSRR246 pKa = 11.84YY247 pKa = 8.66YY248 pKa = 10.32ATGNWVDD255 pKa = 3.47WYY257 pKa = 11.3SLVTIGDD264 pKa = 3.7AGGGASGSGDD274 pKa = 3.79DD275 pKa = 4.27YY276 pKa = 11.82NSGSTQICSTTQEE289 pKa = 4.13RR290 pKa = 11.84KK291 pKa = 9.9NDD293 pKa = 3.26VAEE296 pKa = 4.22NVVSDD301 pKa = 5.01LIEE304 pKa = 4.13STGGVNFGIMRR315 pKa = 11.84FNSSSGGTFMTVSVDD330 pKa = 3.21GQNYY334 pKa = 6.38EE335 pKa = 4.25TTIKK339 pKa = 10.99NMDD342 pKa = 4.45EE343 pKa = 3.8IHH345 pKa = 6.66TGTTTNRR352 pKa = 11.84EE353 pKa = 4.1ALVDD357 pKa = 3.47IASNMPASGYY367 pKa = 8.6TPLGEE372 pKa = 4.29TLYY375 pKa = 11.0EE376 pKa = 3.85AMRR379 pKa = 11.84YY380 pKa = 8.82FSGGNKK386 pKa = 9.18YY387 pKa = 9.91FAGSGSYY394 pKa = 9.29TSPITASCQPNYY406 pKa = 9.85IILMTDD412 pKa = 2.98GMATSDD418 pKa = 3.5SSVPSLSCGGDD429 pKa = 3.54DD430 pKa = 5.91NVDD433 pKa = 3.43CDD435 pKa = 4.04GDD437 pKa = 3.98GRR439 pKa = 11.84SDD441 pKa = 5.31DD442 pKa = 4.75GDD444 pKa = 4.51DD445 pKa = 5.3DD446 pKa = 5.5SLDD449 pKa = 3.63DD450 pKa = 4.31VAWYY454 pKa = 10.01LYY456 pKa = 9.37NTDD459 pKa = 4.52LSDD462 pKa = 4.65FNGTQNVKK470 pKa = 9.62TYY472 pKa = 10.28TIGFGLGGGDD482 pKa = 3.62SDD484 pKa = 6.07AVDD487 pKa = 4.8LLQDD491 pKa = 3.63TADD494 pKa = 3.77NGQGANAGQGKK505 pKa = 10.03SYY507 pKa = 10.38LASSYY512 pKa = 11.2QEE514 pKa = 3.87LTSALSAIIGEE525 pKa = 4.4VLDD528 pKa = 3.68QSSAFVAPVVPTSPEE543 pKa = 3.6NKK545 pKa = 8.99VYY547 pKa = 10.37SGQRR551 pKa = 11.84IYY553 pKa = 11.24LGFFKK558 pKa = 10.62PQITGDD564 pKa = 2.86WYY566 pKa = 11.33GNLKK570 pKa = 10.55KK571 pKa = 10.77FGLDD575 pKa = 3.29NNGEE579 pKa = 4.23VLDD582 pKa = 4.31KK583 pKa = 11.24NGNEE587 pKa = 3.85ATDD590 pKa = 3.65ANGAFLPEE598 pKa = 4.35SVSYY602 pKa = 10.08WGTATDD608 pKa = 3.43AGNVEE613 pKa = 4.27EE614 pKa = 5.05GGIGGILNNRR624 pKa = 11.84VLSTRR629 pKa = 11.84NIYY632 pKa = 9.94TYY634 pKa = 10.59TGTTAALTDD643 pKa = 3.65SSNALTTSNTALTFADD659 pKa = 4.13FDD661 pKa = 3.94VASDD665 pKa = 3.58VRR667 pKa = 11.84KK668 pKa = 10.61DD669 pKa = 3.47EE670 pKa = 4.69IINYY674 pKa = 8.93VYY676 pKa = 10.7GYY678 pKa = 10.34DD679 pKa = 3.34VYY681 pKa = 11.25GINPSVGRR689 pKa = 11.84EE690 pKa = 3.7WVMGDD695 pKa = 3.06ILHH698 pKa = 6.65SKK700 pKa = 9.78PAIQTYY706 pKa = 10.4NSYY709 pKa = 11.15ALSSEE714 pKa = 3.97ADD716 pKa = 3.44PNVNKK721 pKa = 9.58TVIYY725 pKa = 10.43VGSNDD730 pKa = 3.73GQLHH734 pKa = 6.35AFSDD738 pKa = 4.39ANGSEE743 pKa = 4.25LWSFVPPSVLPNLQNLGNNAIHH765 pKa = 7.02EE766 pKa = 4.43YY767 pKa = 11.07FMDD770 pKa = 5.09GSPALYY776 pKa = 10.65VFDD779 pKa = 4.62YY780 pKa = 11.25DD781 pKa = 3.57GDD783 pKa = 4.3GNIGPGPEE791 pKa = 4.64SGDD794 pKa = 3.46TDD796 pKa = 3.68PTGVTDD802 pKa = 5.13AGANDD807 pKa = 3.91KK808 pKa = 11.22VIMVVSMRR816 pKa = 11.84RR817 pKa = 11.84GGGIDD822 pKa = 3.4TLDD825 pKa = 3.61AATSRR830 pKa = 11.84GSYY833 pKa = 9.5YY834 pKa = 11.06ALDD837 pKa = 3.66VTNPLSPQFLWEE849 pKa = 4.12IDD851 pKa = 3.48STTTGFSEE859 pKa = 5.72LGEE862 pKa = 4.3TWSDD866 pKa = 3.11AVIGKK871 pKa = 8.97IRR873 pKa = 11.84LNGSDD878 pKa = 4.54KK879 pKa = 10.5IVAFIGAGYY888 pKa = 10.52DD889 pKa = 3.37NNEE892 pKa = 4.29DD893 pKa = 3.46LRR895 pKa = 11.84FGNTQLFPDD904 pKa = 3.93NTTTATSTILPTADD918 pKa = 3.67ANDD921 pKa = 3.74VTSSGSSAQVNPKK934 pKa = 10.01GRR936 pKa = 11.84GIYY939 pKa = 9.97VIEE942 pKa = 4.58LATVSYY948 pKa = 8.13PTISPFLPTVAVNTSPQKK966 pKa = 9.68LWEE969 pKa = 4.1YY970 pKa = 11.25VYY972 pKa = 11.13DD973 pKa = 4.61ADD975 pKa = 6.28RR976 pKa = 11.84EE977 pKa = 4.17DD978 pKa = 4.49SNLDD982 pKa = 3.44AYY984 pKa = 11.0DD985 pKa = 4.0PGNNPKK991 pKa = 10.12YY992 pKa = 10.9SFVATIATLDD1002 pKa = 3.7SDD1004 pKa = 3.69FDD1006 pKa = 4.39GYY1008 pKa = 10.54IDD1010 pKa = 4.01RR1011 pKa = 11.84LYY1013 pKa = 11.31SGDD1016 pKa = 3.19TGGNMWRR1023 pKa = 11.84FDD1025 pKa = 3.66IGDD1028 pKa = 3.83KK1029 pKa = 10.44TSTSAWDD1036 pKa = 3.26ATKK1039 pKa = 10.33IFSANPSDD1047 pKa = 3.98VANSDD1052 pKa = 3.68EE1053 pKa = 4.96DD1054 pKa = 3.88PATNGRR1060 pKa = 11.84KK1061 pKa = 8.97IFYY1064 pKa = 9.69RR1065 pKa = 11.84PSIVQEE1071 pKa = 3.75PGYY1074 pKa = 10.27VGVYY1078 pKa = 10.29FGTGDD1083 pKa = 3.7RR1084 pKa = 11.84AHH1086 pKa = 7.5PLNKK1090 pKa = 10.09AVIDD1094 pKa = 3.82RR1095 pKa = 11.84LYY1097 pKa = 11.19AVYY1100 pKa = 10.54DD1101 pKa = 3.6RR1102 pKa = 11.84GNITSARR1109 pKa = 11.84TEE1111 pKa = 3.79EE1112 pKa = 4.18HH1113 pKa = 6.13LVNVTEE1119 pKa = 5.89DD1120 pKa = 3.73NLQSASPGIGVVNSTLLALSSTSNYY1145 pKa = 9.75GWFIKK1150 pKa = 10.59LDD1152 pKa = 3.84LLDD1155 pKa = 4.14GEE1157 pKa = 5.2KK1158 pKa = 10.72VLSNPIVFNKK1168 pKa = 8.58VAYY1171 pKa = 7.71YY1172 pKa = 7.36TTFTANVPSDD1182 pKa = 3.99DD1183 pKa = 3.98PCEE1186 pKa = 3.85QGQLGIGRR1194 pKa = 11.84LYY1196 pKa = 11.01AVDD1199 pKa = 4.01YY1200 pKa = 8.34KK1201 pKa = 10.91TGEE1204 pKa = 3.87AVFNFDD1210 pKa = 4.13LTNDD1214 pKa = 3.45VADD1217 pKa = 3.89EE1218 pKa = 4.15DD1219 pKa = 4.98SYY1221 pKa = 12.26AEE1223 pKa = 4.13NEE1225 pKa = 4.12NEE1227 pKa = 3.82RR1228 pKa = 11.84AIAGGGKK1235 pKa = 9.14ILRR1238 pKa = 11.84RR1239 pKa = 11.84SDD1241 pKa = 3.2RR1242 pKa = 11.84VKK1244 pKa = 10.21TIGSGIPSGAVVVVRR1259 pKa = 11.84EE1260 pKa = 4.78DD1261 pKa = 3.6GTSSALIGCGGGLCGGEE1278 pKa = 4.14TKK1280 pKa = 10.81GGGTAVPIYY1289 pKa = 10.74WMMEE1293 pKa = 3.59
Molecular weight: 138.13 kDa
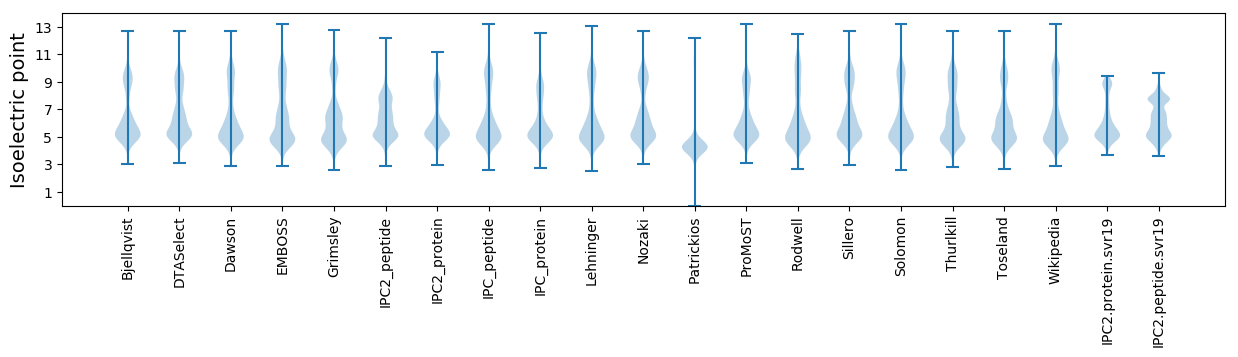
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6JW85|A0A1M6JW85_MALRU Uncharacterized protein OS=Malonomonas rubra DSM 5091 OX=1122189 GN=SAMN02745165_02541 PE=4 SV=1
MM1 pKa = 7.55SKK3 pKa = 10.43PKK5 pKa = 9.39RR6 pKa = 11.84TYY8 pKa = 8.97QPSRR12 pKa = 11.84IKK14 pKa = 10.52RR15 pKa = 11.84KK16 pKa = 8.04RR17 pKa = 11.84THH19 pKa = 6.13GFRR22 pKa = 11.84KK23 pKa = 10.09RR24 pKa = 11.84MQTKK28 pKa = 10.03SGRR31 pKa = 11.84LVINRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84SRR40 pKa = 11.84GRR42 pKa = 11.84KK43 pKa = 8.35SLSVTTPQKK52 pKa = 11.05
MM1 pKa = 7.55SKK3 pKa = 10.43PKK5 pKa = 9.39RR6 pKa = 11.84TYY8 pKa = 8.97QPSRR12 pKa = 11.84IKK14 pKa = 10.52RR15 pKa = 11.84KK16 pKa = 8.04RR17 pKa = 11.84THH19 pKa = 6.13GFRR22 pKa = 11.84KK23 pKa = 10.09RR24 pKa = 11.84MQTKK28 pKa = 10.03SGRR31 pKa = 11.84LVINRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84SRR40 pKa = 11.84GRR42 pKa = 11.84KK43 pKa = 8.35SLSVTTPQKK52 pKa = 11.05
Molecular weight: 6.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1195547 |
40 |
4834 |
327.2 |
36.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.793 ± 0.059 | 1.263 ± 0.022 |
5.597 ± 0.046 | 6.909 ± 0.042 |
4.106 ± 0.033 | 7.427 ± 0.045 |
2.003 ± 0.024 | 6.004 ± 0.04 |
5.185 ± 0.039 | 11.155 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.46 ± 0.024 | 3.485 ± 0.029 |
4.254 ± 0.029 | 4.45 ± 0.04 |
5.433 ± 0.041 | 5.999 ± 0.047 |
4.817 ± 0.039 | 6.854 ± 0.032 |
1.04 ± 0.014 | 2.766 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
