
Proteus phage PM 85
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Molineuxvirinae; Acadevirus; Proteus virus PM85
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
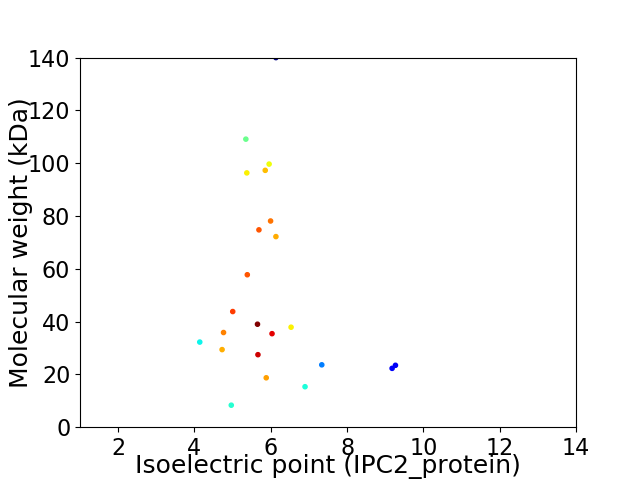
Virtual 2D-PAGE plot for 23 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
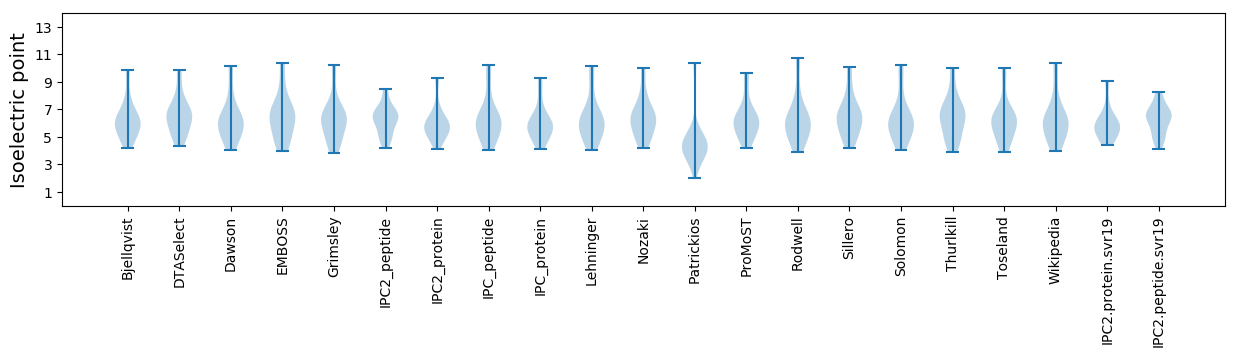
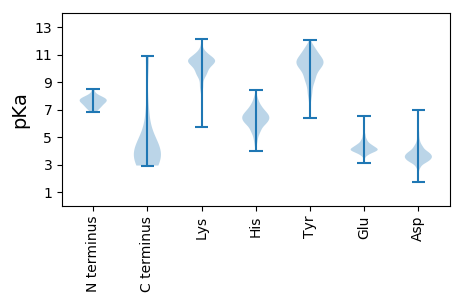
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6NYA6|A0A0F6NYA6_9CAUD Uncharacterized protein OS=Proteus phage PM 85 OX=1560283 GN=PM85_009 PE=4 SV=1
MM1 pKa = 7.51AFEE4 pKa = 4.52IVDD7 pKa = 3.8TEE9 pKa = 4.55TTQDD13 pKa = 3.02TSTTTEE19 pKa = 3.39VDD21 pKa = 3.33TQEE24 pKa = 4.44VKK26 pKa = 10.79DD27 pKa = 4.06VNNNDD32 pKa = 3.41NTDD35 pKa = 3.38TGSTDD40 pKa = 3.83DD41 pKa = 3.82VQGNPAPEE49 pKa = 4.38GDD51 pKa = 3.98EE52 pKa = 4.03NSGGEE57 pKa = 4.05DD58 pKa = 2.87TGGNGQDD65 pKa = 5.11DD66 pKa = 4.0KK67 pKa = 11.75QEE69 pKa = 4.15PQGTEE74 pKa = 3.28DD75 pKa = 2.86STYY78 pKa = 9.7YY79 pKa = 10.68FGGNEE84 pKa = 3.97VEE86 pKa = 5.01IEE88 pKa = 3.94IPQDD92 pKa = 3.35VEE94 pKa = 3.88EE95 pKa = 4.28SLKK98 pKa = 11.07EE99 pKa = 3.8KK100 pKa = 10.94GIDD103 pKa = 3.37AKK105 pKa = 11.06EE106 pKa = 3.88LVAEE110 pKa = 5.19LYY112 pKa = 11.06SKK114 pKa = 11.21DD115 pKa = 3.46GDD117 pKa = 3.67FSLSDD122 pKa = 3.23EE123 pKa = 4.73TKK125 pKa = 10.55QKK127 pKa = 11.03LYY129 pKa = 11.18DD130 pKa = 3.63AFGKK134 pKa = 10.22FAVDD138 pKa = 4.34AYY140 pKa = 11.39LSGLKK145 pKa = 9.87AQNEE149 pKa = 4.3MFFMNEE155 pKa = 3.17ATKK158 pKa = 10.64AKK160 pKa = 10.33EE161 pKa = 3.88MEE163 pKa = 4.04QANAQRR169 pKa = 11.84FNDD172 pKa = 3.24IAKK175 pKa = 10.16EE176 pKa = 4.04VGGDD180 pKa = 3.66EE181 pKa = 3.93GWTRR185 pKa = 11.84LEE187 pKa = 3.93EE188 pKa = 4.25FALSTLSNEE197 pKa = 3.9EE198 pKa = 3.66LTAFNAVMDD207 pKa = 4.39SGNQYY212 pKa = 10.08LQMYY216 pKa = 9.16AVRR219 pKa = 11.84EE220 pKa = 3.95LEE222 pKa = 3.91ARR224 pKa = 11.84RR225 pKa = 11.84KK226 pKa = 8.36QAQGDD231 pKa = 4.02DD232 pKa = 3.77KK233 pKa = 11.86VEE235 pKa = 4.04LVQGTPASDD244 pKa = 3.55EE245 pKa = 4.38GDD247 pKa = 3.55NSPLSAQEE255 pKa = 4.01YY256 pKa = 8.47IRR258 pKa = 11.84EE259 pKa = 4.07VAQIGSKK266 pKa = 10.47FRR268 pKa = 11.84GDD270 pKa = 2.9RR271 pKa = 11.84KK272 pKa = 10.44GAAEE276 pKa = 4.07YY277 pKa = 9.7QAKK280 pKa = 10.38LDD282 pKa = 3.46ARR284 pKa = 11.84RR285 pKa = 11.84RR286 pKa = 11.84AGMARR291 pKa = 11.84GLL293 pKa = 3.78
MM1 pKa = 7.51AFEE4 pKa = 4.52IVDD7 pKa = 3.8TEE9 pKa = 4.55TTQDD13 pKa = 3.02TSTTTEE19 pKa = 3.39VDD21 pKa = 3.33TQEE24 pKa = 4.44VKK26 pKa = 10.79DD27 pKa = 4.06VNNNDD32 pKa = 3.41NTDD35 pKa = 3.38TGSTDD40 pKa = 3.83DD41 pKa = 3.82VQGNPAPEE49 pKa = 4.38GDD51 pKa = 3.98EE52 pKa = 4.03NSGGEE57 pKa = 4.05DD58 pKa = 2.87TGGNGQDD65 pKa = 5.11DD66 pKa = 4.0KK67 pKa = 11.75QEE69 pKa = 4.15PQGTEE74 pKa = 3.28DD75 pKa = 2.86STYY78 pKa = 9.7YY79 pKa = 10.68FGGNEE84 pKa = 3.97VEE86 pKa = 5.01IEE88 pKa = 3.94IPQDD92 pKa = 3.35VEE94 pKa = 3.88EE95 pKa = 4.28SLKK98 pKa = 11.07EE99 pKa = 3.8KK100 pKa = 10.94GIDD103 pKa = 3.37AKK105 pKa = 11.06EE106 pKa = 3.88LVAEE110 pKa = 5.19LYY112 pKa = 11.06SKK114 pKa = 11.21DD115 pKa = 3.46GDD117 pKa = 3.67FSLSDD122 pKa = 3.23EE123 pKa = 4.73TKK125 pKa = 10.55QKK127 pKa = 11.03LYY129 pKa = 11.18DD130 pKa = 3.63AFGKK134 pKa = 10.22FAVDD138 pKa = 4.34AYY140 pKa = 11.39LSGLKK145 pKa = 9.87AQNEE149 pKa = 4.3MFFMNEE155 pKa = 3.17ATKK158 pKa = 10.64AKK160 pKa = 10.33EE161 pKa = 3.88MEE163 pKa = 4.04QANAQRR169 pKa = 11.84FNDD172 pKa = 3.24IAKK175 pKa = 10.16EE176 pKa = 4.04VGGDD180 pKa = 3.66EE181 pKa = 3.93GWTRR185 pKa = 11.84LEE187 pKa = 3.93EE188 pKa = 4.25FALSTLSNEE197 pKa = 3.9EE198 pKa = 3.66LTAFNAVMDD207 pKa = 4.39SGNQYY212 pKa = 10.08LQMYY216 pKa = 9.16AVRR219 pKa = 11.84EE220 pKa = 3.95LEE222 pKa = 3.91ARR224 pKa = 11.84RR225 pKa = 11.84KK226 pKa = 8.36QAQGDD231 pKa = 4.02DD232 pKa = 3.77KK233 pKa = 11.86VEE235 pKa = 4.04LVQGTPASDD244 pKa = 3.55EE245 pKa = 4.38GDD247 pKa = 3.55NSPLSAQEE255 pKa = 4.01YY256 pKa = 8.47IRR258 pKa = 11.84EE259 pKa = 4.07VAQIGSKK266 pKa = 10.47FRR268 pKa = 11.84GDD270 pKa = 2.9RR271 pKa = 11.84KK272 pKa = 10.44GAAEE276 pKa = 4.07YY277 pKa = 9.7QAKK280 pKa = 10.38LDD282 pKa = 3.46ARR284 pKa = 11.84RR285 pKa = 11.84RR286 pKa = 11.84AGMARR291 pKa = 11.84GLL293 pKa = 3.78
Molecular weight: 32.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6NYI6|A0A0F6NYI6_9CAUD DNA ligase OS=Proteus phage PM 85 OX=1560283 GN=PM85_0010 PE=4 SV=1
MM1 pKa = 6.98YY2 pKa = 9.98QLKK5 pKa = 10.68VNIGSRR11 pKa = 11.84VRR13 pKa = 11.84NIHH16 pKa = 5.47KK17 pKa = 10.35HH18 pKa = 3.97SVNYY22 pKa = 9.73GKK24 pKa = 10.47SGFIEE29 pKa = 4.56RR30 pKa = 11.84EE31 pKa = 3.98VLGVKK36 pKa = 9.59PSKK39 pKa = 10.18WLVRR43 pKa = 11.84YY44 pKa = 9.96NDD46 pKa = 4.28GSCGTYY52 pKa = 10.26FKK54 pKa = 10.0HH55 pKa = 5.45TAHH58 pKa = 7.55RR59 pKa = 11.84SLQVVDD65 pKa = 4.62EE66 pKa = 4.42YY67 pKa = 11.71KK68 pKa = 10.58SVKK71 pKa = 8.11QTIKK75 pKa = 10.04EE76 pKa = 3.99QQKK79 pKa = 9.22EE80 pKa = 3.94EE81 pKa = 4.45RR82 pKa = 11.84IMIKK86 pKa = 9.77RR87 pKa = 11.84EE88 pKa = 3.49RR89 pKa = 11.84RR90 pKa = 11.84NVITGWTQSEE100 pKa = 4.06LMKK103 pKa = 10.61RR104 pKa = 11.84HH105 pKa = 5.22GNAMGVVQFNQRR117 pKa = 11.84ALGKK121 pKa = 7.47TTGQAMAVIGSAMVQPYY138 pKa = 10.02VPVSFHH144 pKa = 6.81NIDD147 pKa = 3.33HH148 pKa = 7.35AIFEE152 pKa = 5.11HH153 pKa = 7.05NIPTQVANANMRR165 pKa = 11.84DD166 pKa = 3.82MIKK169 pKa = 10.86DD170 pKa = 4.29LINTLGFKK178 pKa = 10.44GFRR181 pKa = 11.84FNNNNNTIVFNPIVTEE197 pKa = 3.97EE198 pKa = 4.35NYY200 pKa = 10.27VEE202 pKa = 4.6SVV204 pKa = 2.93
MM1 pKa = 6.98YY2 pKa = 9.98QLKK5 pKa = 10.68VNIGSRR11 pKa = 11.84VRR13 pKa = 11.84NIHH16 pKa = 5.47KK17 pKa = 10.35HH18 pKa = 3.97SVNYY22 pKa = 9.73GKK24 pKa = 10.47SGFIEE29 pKa = 4.56RR30 pKa = 11.84EE31 pKa = 3.98VLGVKK36 pKa = 9.59PSKK39 pKa = 10.18WLVRR43 pKa = 11.84YY44 pKa = 9.96NDD46 pKa = 4.28GSCGTYY52 pKa = 10.26FKK54 pKa = 10.0HH55 pKa = 5.45TAHH58 pKa = 7.55RR59 pKa = 11.84SLQVVDD65 pKa = 4.62EE66 pKa = 4.42YY67 pKa = 11.71KK68 pKa = 10.58SVKK71 pKa = 8.11QTIKK75 pKa = 10.04EE76 pKa = 3.99QQKK79 pKa = 9.22EE80 pKa = 3.94EE81 pKa = 4.45RR82 pKa = 11.84IMIKK86 pKa = 9.77RR87 pKa = 11.84EE88 pKa = 3.49RR89 pKa = 11.84RR90 pKa = 11.84NVITGWTQSEE100 pKa = 4.06LMKK103 pKa = 10.61RR104 pKa = 11.84HH105 pKa = 5.22GNAMGVVQFNQRR117 pKa = 11.84ALGKK121 pKa = 7.47TTGQAMAVIGSAMVQPYY138 pKa = 10.02VPVSFHH144 pKa = 6.81NIDD147 pKa = 3.33HH148 pKa = 7.35AIFEE152 pKa = 5.11HH153 pKa = 7.05NIPTQVANANMRR165 pKa = 11.84DD166 pKa = 3.82MIKK169 pKa = 10.86DD170 pKa = 4.29LINTLGFKK178 pKa = 10.44GFRR181 pKa = 11.84FNNNNNTIVFNPIVTEE197 pKa = 3.97EE198 pKa = 4.35NYY200 pKa = 10.27VEE202 pKa = 4.6SVV204 pKa = 2.93
Molecular weight: 23.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10872 |
72 |
1273 |
472.7 |
52.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.864 ± 0.554 | 0.92 ± 0.184 |
6.429 ± 0.252 | 7.009 ± 0.352 |
3.909 ± 0.159 | 7.791 ± 0.294 |
1.904 ± 0.244 | 5.85 ± 0.278 |
6.714 ± 0.267 | 7.745 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.164 ± 0.224 | 4.746 ± 0.263 |
3.339 ± 0.18 | 4.295 ± 0.289 |
5.362 ± 0.384 | 6.025 ± 0.38 |
5.427 ± 0.441 | 6.751 ± 0.35 |
1.38 ± 0.147 | 3.376 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
