
Candidatus [Bacteroides] periocalifornicus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidetes incertae sedis
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
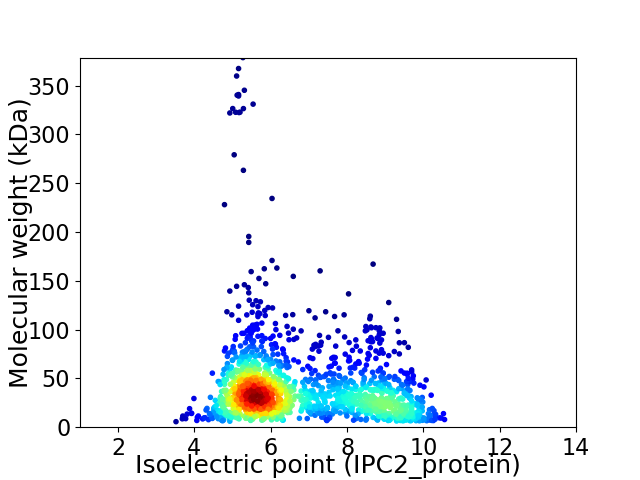
Virtual 2D-PAGE plot for 1579 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q4B934|A0A0Q4B934_9BACT Uncharacterized protein OS=Candidatus [Bacteroides] periocalifornicus OX=1702214 GN=AL399_02645 PE=4 SV=1
MM1 pKa = 6.95NTLKK5 pKa = 10.75GLYY8 pKa = 9.16PDD10 pKa = 4.4YY11 pKa = 10.64EE12 pKa = 4.46PNIAFYY18 pKa = 10.3NAEE21 pKa = 4.14VEE23 pKa = 4.67DD24 pKa = 4.59EE25 pKa = 4.38PACVMVDD32 pKa = 3.9LNLEE36 pKa = 3.87EE37 pKa = 4.23YY38 pKa = 10.91APVEE42 pKa = 4.22GYY44 pKa = 10.75EE45 pKa = 4.19YY46 pKa = 10.19ICVVKK51 pKa = 10.61VLAPEE56 pKa = 4.45GASINGDD63 pKa = 3.37KK64 pKa = 10.49IGRR67 pKa = 11.84LDD69 pKa = 3.71EE70 pKa = 4.63KK71 pKa = 10.44LTEE74 pKa = 4.52HH75 pKa = 6.94LAEE78 pKa = 4.88AYY80 pKa = 9.5GAVWAGSLNWDD91 pKa = 3.78DD92 pKa = 4.23EE93 pKa = 4.55VHH95 pKa = 5.71IFYY98 pKa = 10.95YY99 pKa = 9.24LTEE102 pKa = 4.94EE103 pKa = 3.85IDD105 pKa = 3.34QRR107 pKa = 11.84EE108 pKa = 4.24ILQFPPVKK116 pKa = 9.96AAKK119 pKa = 9.24YY120 pKa = 9.36DD121 pKa = 4.24CIVEE125 pKa = 4.24IAPEE129 pKa = 4.36PEE131 pKa = 3.44WDD133 pKa = 3.42TYY135 pKa = 11.51LALLYY140 pKa = 10.02PDD142 pKa = 4.61DD143 pKa = 4.12YY144 pKa = 11.88GRR146 pKa = 11.84LEE148 pKa = 3.81IEE150 pKa = 3.82NRR152 pKa = 11.84EE153 pKa = 4.17LLGEE157 pKa = 4.16AQGEE161 pKa = 4.18GDD163 pKa = 3.75DD164 pKa = 3.95FSRR167 pKa = 11.84ARR169 pKa = 11.84LVTHH173 pKa = 6.4SSYY176 pKa = 11.13FDD178 pKa = 3.3RR179 pKa = 11.84QEE181 pKa = 3.93DD182 pKa = 3.61LKK184 pKa = 11.41AFCTEE189 pKa = 3.81VEE191 pKa = 4.03RR192 pKa = 11.84LGFRR196 pKa = 11.84IVEE199 pKa = 4.28LEE201 pKa = 4.02DD202 pKa = 3.89TPDD205 pKa = 3.04VDD207 pKa = 5.02ADD209 pKa = 3.77FPYY212 pKa = 9.56GVEE215 pKa = 3.96YY216 pKa = 11.06AKK218 pKa = 10.77VHH220 pKa = 5.7TLLDD224 pKa = 3.5IPKK227 pKa = 8.36ITQEE231 pKa = 4.08LLEE234 pKa = 4.69LTLAHH239 pKa = 6.39NGYY242 pKa = 10.48YY243 pKa = 10.34DD244 pKa = 3.23GWLNTSDD251 pKa = 5.64ADD253 pKa = 4.78IDD255 pKa = 4.02DD256 pKa = 4.07LL257 pKa = 5.61
MM1 pKa = 6.95NTLKK5 pKa = 10.75GLYY8 pKa = 9.16PDD10 pKa = 4.4YY11 pKa = 10.64EE12 pKa = 4.46PNIAFYY18 pKa = 10.3NAEE21 pKa = 4.14VEE23 pKa = 4.67DD24 pKa = 4.59EE25 pKa = 4.38PACVMVDD32 pKa = 3.9LNLEE36 pKa = 3.87EE37 pKa = 4.23YY38 pKa = 10.91APVEE42 pKa = 4.22GYY44 pKa = 10.75EE45 pKa = 4.19YY46 pKa = 10.19ICVVKK51 pKa = 10.61VLAPEE56 pKa = 4.45GASINGDD63 pKa = 3.37KK64 pKa = 10.49IGRR67 pKa = 11.84LDD69 pKa = 3.71EE70 pKa = 4.63KK71 pKa = 10.44LTEE74 pKa = 4.52HH75 pKa = 6.94LAEE78 pKa = 4.88AYY80 pKa = 9.5GAVWAGSLNWDD91 pKa = 3.78DD92 pKa = 4.23EE93 pKa = 4.55VHH95 pKa = 5.71IFYY98 pKa = 10.95YY99 pKa = 9.24LTEE102 pKa = 4.94EE103 pKa = 3.85IDD105 pKa = 3.34QRR107 pKa = 11.84EE108 pKa = 4.24ILQFPPVKK116 pKa = 9.96AAKK119 pKa = 9.24YY120 pKa = 9.36DD121 pKa = 4.24CIVEE125 pKa = 4.24IAPEE129 pKa = 4.36PEE131 pKa = 3.44WDD133 pKa = 3.42TYY135 pKa = 11.51LALLYY140 pKa = 10.02PDD142 pKa = 4.61DD143 pKa = 4.12YY144 pKa = 11.88GRR146 pKa = 11.84LEE148 pKa = 3.81IEE150 pKa = 3.82NRR152 pKa = 11.84EE153 pKa = 4.17LLGEE157 pKa = 4.16AQGEE161 pKa = 4.18GDD163 pKa = 3.75DD164 pKa = 3.95FSRR167 pKa = 11.84ARR169 pKa = 11.84LVTHH173 pKa = 6.4SSYY176 pKa = 11.13FDD178 pKa = 3.3RR179 pKa = 11.84QEE181 pKa = 3.93DD182 pKa = 3.61LKK184 pKa = 11.41AFCTEE189 pKa = 3.81VEE191 pKa = 4.03RR192 pKa = 11.84LGFRR196 pKa = 11.84IVEE199 pKa = 4.28LEE201 pKa = 4.02DD202 pKa = 3.89TPDD205 pKa = 3.04VDD207 pKa = 5.02ADD209 pKa = 3.77FPYY212 pKa = 9.56GVEE215 pKa = 3.96YY216 pKa = 11.06AKK218 pKa = 10.77VHH220 pKa = 5.7TLLDD224 pKa = 3.5IPKK227 pKa = 8.36ITQEE231 pKa = 4.08LLEE234 pKa = 4.69LTLAHH239 pKa = 6.39NGYY242 pKa = 10.48YY243 pKa = 10.34DD244 pKa = 3.23GWLNTSDD251 pKa = 5.64ADD253 pKa = 4.78IDD255 pKa = 4.02DD256 pKa = 4.07LL257 pKa = 5.61
Molecular weight: 29.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q4B339|A0A0Q4B339_9BACT Nitrogen fixation protein NifU OS=Candidatus [Bacteroides] periocalifornicus OX=1702214 GN=AL399_07790 PE=4 SV=1
MM1 pKa = 7.85AEE3 pKa = 3.62QIARR7 pKa = 11.84IRR9 pKa = 11.84LIHH12 pKa = 4.95TAKK15 pKa = 10.42VGEE18 pKa = 4.21KK19 pKa = 10.57FEE21 pKa = 4.27IASVRR26 pKa = 11.84GLGSLTRR33 pKa = 11.84RR34 pKa = 11.84GIARR38 pKa = 11.84SSKK41 pKa = 10.26GGHH44 pKa = 5.38TLRR47 pKa = 11.84SASPLCDD54 pKa = 3.35NMDD57 pKa = 5.33GIHH60 pKa = 6.23LWWRR64 pKa = 11.84EE65 pKa = 3.75NLRR68 pKa = 11.84VAKK71 pKa = 7.9MVCVYY76 pKa = 10.75GEE78 pKa = 3.73PDD80 pKa = 2.85GRR82 pKa = 11.84GNLRR86 pKa = 11.84VAKK89 pKa = 9.2TPPLQSALAGMLLLVALGVVDD110 pKa = 5.68KK111 pKa = 11.37INVFCGGLPMQGRR124 pKa = 11.84GAPRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 10.35KK131 pKa = 10.89NGGDD135 pKa = 3.07HH136 pKa = 6.95ALNRR140 pKa = 11.84EE141 pKa = 3.9GGLIHH146 pKa = 7.5RR147 pKa = 11.84DD148 pKa = 3.29AGNSS152 pKa = 3.32
MM1 pKa = 7.85AEE3 pKa = 3.62QIARR7 pKa = 11.84IRR9 pKa = 11.84LIHH12 pKa = 4.95TAKK15 pKa = 10.42VGEE18 pKa = 4.21KK19 pKa = 10.57FEE21 pKa = 4.27IASVRR26 pKa = 11.84GLGSLTRR33 pKa = 11.84RR34 pKa = 11.84GIARR38 pKa = 11.84SSKK41 pKa = 10.26GGHH44 pKa = 5.38TLRR47 pKa = 11.84SASPLCDD54 pKa = 3.35NMDD57 pKa = 5.33GIHH60 pKa = 6.23LWWRR64 pKa = 11.84EE65 pKa = 3.75NLRR68 pKa = 11.84VAKK71 pKa = 7.9MVCVYY76 pKa = 10.75GEE78 pKa = 3.73PDD80 pKa = 2.85GRR82 pKa = 11.84GNLRR86 pKa = 11.84VAKK89 pKa = 9.2TPPLQSALAGMLLLVALGVVDD110 pKa = 5.68KK111 pKa = 11.37INVFCGGLPMQGRR124 pKa = 11.84GAPRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 10.35KK131 pKa = 10.89NGGDD135 pKa = 3.07HH136 pKa = 6.95ALNRR140 pKa = 11.84EE141 pKa = 3.9GGLIHH146 pKa = 7.5RR147 pKa = 11.84DD148 pKa = 3.29AGNSS152 pKa = 3.32
Molecular weight: 16.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605109 |
56 |
3444 |
383.2 |
42.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.151 ± 0.05 | 1.063 ± 0.02 |
4.882 ± 0.04 | 6.266 ± 0.053 |
3.782 ± 0.037 | 8.148 ± 0.055 |
2.023 ± 0.027 | 5.097 ± 0.052 |
4.324 ± 0.055 | 10.409 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.033 | 3.553 ± 0.04 |
4.638 ± 0.038 | 4.049 ± 0.034 |
6.268 ± 0.066 | 5.846 ± 0.05 |
5.532 ± 0.07 | 7.648 ± 0.055 |
1.256 ± 0.022 | 3.776 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
