
Sida yellow vein Madurai virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.32
Get precalculated fractions of proteins
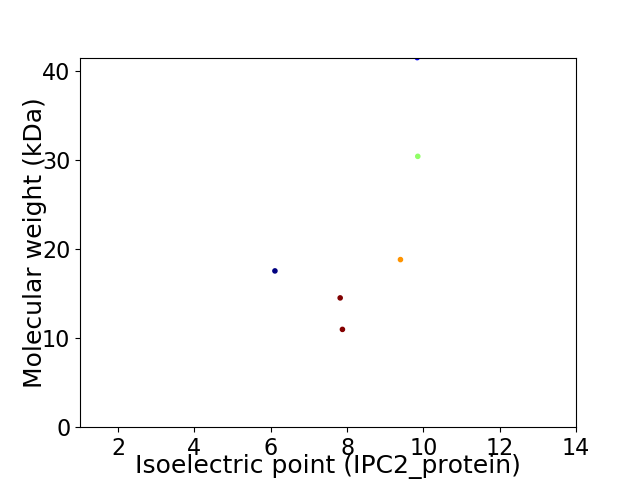
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q17ZR2|Q17ZR2_9GEMI V2 protein OS=Sida yellow vein Madurai virus OX=420255 GN=v2 PE=3 SV=1
MM1 pKa = 7.98WDD3 pKa = 3.34PLEE6 pKa = 4.57NEE8 pKa = 4.32FPEE11 pKa = 4.69TVHH14 pKa = 6.36GFRR17 pKa = 11.84YY18 pKa = 8.53MLAVKK23 pKa = 9.92YY24 pKa = 7.84MQEE27 pKa = 3.89IAKK30 pKa = 8.53STSLVHH36 pKa = 5.96WFRR39 pKa = 11.84VRR41 pKa = 11.84SGSHH45 pKa = 5.26FRR47 pKa = 11.84VEE49 pKa = 4.29VQGLCSSVLQVWRR62 pKa = 11.84FPLQFLPRR70 pKa = 11.84VEE72 pKa = 4.77GSTSTVLAHH81 pKa = 6.97QLLLPLLPQAQGEE94 pKa = 4.38EE95 pKa = 4.27DD96 pKa = 4.85GPTGPCIEE104 pKa = 4.73SPDD107 pKa = 3.46FTEE110 pKa = 4.92CIEE113 pKa = 4.12RR114 pKa = 11.84LMSLRR119 pKa = 11.84AVKK122 pKa = 10.48AHH124 pKa = 5.13VRR126 pKa = 11.84YY127 pKa = 9.69SPSMLKK133 pKa = 9.86TILVTWVRR141 pKa = 11.84LSVYY145 pKa = 10.9LMLLEE150 pKa = 4.13VLGG153 pKa = 4.19
MM1 pKa = 7.98WDD3 pKa = 3.34PLEE6 pKa = 4.57NEE8 pKa = 4.32FPEE11 pKa = 4.69TVHH14 pKa = 6.36GFRR17 pKa = 11.84YY18 pKa = 8.53MLAVKK23 pKa = 9.92YY24 pKa = 7.84MQEE27 pKa = 3.89IAKK30 pKa = 8.53STSLVHH36 pKa = 5.96WFRR39 pKa = 11.84VRR41 pKa = 11.84SGSHH45 pKa = 5.26FRR47 pKa = 11.84VEE49 pKa = 4.29VQGLCSSVLQVWRR62 pKa = 11.84FPLQFLPRR70 pKa = 11.84VEE72 pKa = 4.77GSTSTVLAHH81 pKa = 6.97QLLLPLLPQAQGEE94 pKa = 4.38EE95 pKa = 4.27DD96 pKa = 4.85GPTGPCIEE104 pKa = 4.73SPDD107 pKa = 3.46FTEE110 pKa = 4.92CIEE113 pKa = 4.12RR114 pKa = 11.84LMSLRR119 pKa = 11.84AVKK122 pKa = 10.48AHH124 pKa = 5.13VRR126 pKa = 11.84YY127 pKa = 9.69SPSMLKK133 pKa = 9.86TILVTWVRR141 pKa = 11.84LSVYY145 pKa = 10.9LMLLEE150 pKa = 4.13VLGG153 pKa = 4.19
Molecular weight: 17.53 kDa
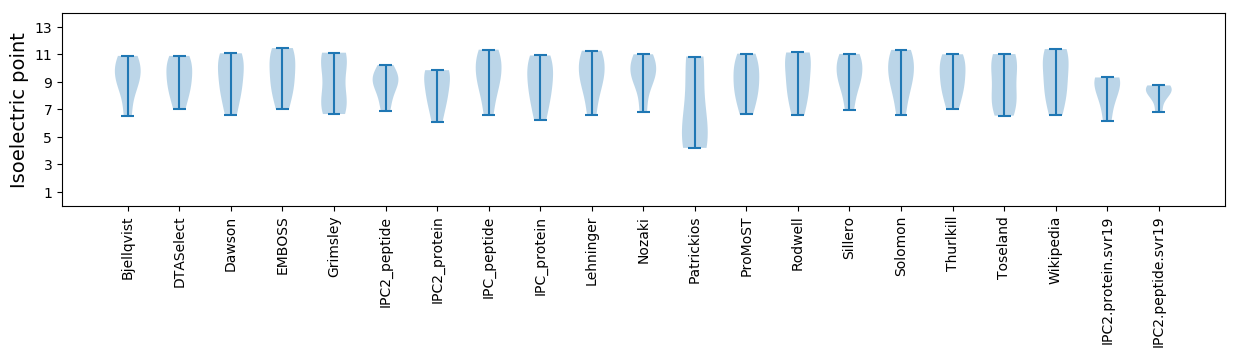
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q17ZQ9|Q17ZQ9_9GEMI Transcriptional activator protein OS=Sida yellow vein Madurai virus OX=420255 GN=c2 PE=3 SV=1
MM1 pKa = 7.43APSKK5 pKa = 10.57RR6 pKa = 11.84FNIYY10 pKa = 9.69CKK12 pKa = 10.48NYY14 pKa = 8.63FLTYY18 pKa = 9.03PKK20 pKa = 10.53CSLTKK25 pKa = 10.66EE26 pKa = 4.11EE27 pKa = 5.14ALSQIQNLQTPTNKK41 pKa = 10.17KK42 pKa = 9.08YY43 pKa = 10.8IKK45 pKa = 9.86VCKK48 pKa = 9.17EE49 pKa = 3.37LHH51 pKa = 6.47EE52 pKa = 4.83NGEE55 pKa = 4.16PHH57 pKa = 6.66LHH59 pKa = 6.03VLIQFEE65 pKa = 4.78GKK67 pKa = 9.95YY68 pKa = 9.77KK69 pKa = 10.46CQNQRR74 pKa = 11.84FFDD77 pKa = 3.9LVSPNRR83 pKa = 11.84SAHH86 pKa = 5.06FHH88 pKa = 6.56PNIQGAKK95 pKa = 9.25SSSDD99 pKa = 3.11VKK101 pKa = 11.24SYY103 pKa = 10.79IDD105 pKa = 3.7KK106 pKa = 11.33DD107 pKa = 3.49GDD109 pKa = 3.65TLEE112 pKa = 4.14WGEE115 pKa = 3.84IQIDD119 pKa = 3.71GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 4.41AYY134 pKa = 9.38PPQRR138 pKa = 11.84FTQAVSQRR146 pKa = 11.84LLQYY150 pKa = 10.83LEE152 pKa = 4.55NYY154 pKa = 8.24PLRR157 pKa = 11.84IMFYY161 pKa = 10.62NFIIYY166 pKa = 8.54MLIYY170 pKa = 10.23IGFSHH175 pKa = 7.13LLWRR179 pKa = 11.84FIFLLFLLLLSIKK192 pKa = 10.35FPTNLKK198 pKa = 9.39SGRR201 pKa = 11.84VRR203 pKa = 11.84MSWRR207 pKa = 11.84PLRR210 pKa = 11.84GLGGPSVLWLKK221 pKa = 9.1VTVEE225 pKa = 3.97RR226 pKa = 11.84GRR228 pKa = 11.84RR229 pKa = 11.84CGPGHH234 pKa = 6.43WVHH237 pKa = 7.9IIISVGIWTSAQRR250 pKa = 11.84YY251 pKa = 5.06TVTTLGTTSLMTLIPIISNTLKK273 pKa = 10.86SLWGPRR279 pKa = 11.84EE280 pKa = 3.81TGKK283 pKa = 10.62AIPSTASQFKK293 pKa = 10.44LKK295 pKa = 10.77AGFQQSSFAIQGPIPAIKK313 pKa = 10.58NSWTRR318 pKa = 11.84KK319 pKa = 9.19RR320 pKa = 11.84IPLSRR325 pKa = 11.84PGHH328 pKa = 5.5FKK330 pKa = 10.77CVVRR334 pKa = 11.84YY335 pKa = 7.95PRR337 pKa = 11.84RR338 pKa = 11.84GSLLRR343 pKa = 11.84CPSKK347 pKa = 10.69SSIRR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84RR354 pKa = 11.84GKK356 pKa = 10.06YY357 pKa = 9.64AGRR360 pKa = 11.84GG361 pKa = 3.46
MM1 pKa = 7.43APSKK5 pKa = 10.57RR6 pKa = 11.84FNIYY10 pKa = 9.69CKK12 pKa = 10.48NYY14 pKa = 8.63FLTYY18 pKa = 9.03PKK20 pKa = 10.53CSLTKK25 pKa = 10.66EE26 pKa = 4.11EE27 pKa = 5.14ALSQIQNLQTPTNKK41 pKa = 10.17KK42 pKa = 9.08YY43 pKa = 10.8IKK45 pKa = 9.86VCKK48 pKa = 9.17EE49 pKa = 3.37LHH51 pKa = 6.47EE52 pKa = 4.83NGEE55 pKa = 4.16PHH57 pKa = 6.66LHH59 pKa = 6.03VLIQFEE65 pKa = 4.78GKK67 pKa = 9.95YY68 pKa = 9.77KK69 pKa = 10.46CQNQRR74 pKa = 11.84FFDD77 pKa = 3.9LVSPNRR83 pKa = 11.84SAHH86 pKa = 5.06FHH88 pKa = 6.56PNIQGAKK95 pKa = 9.25SSSDD99 pKa = 3.11VKK101 pKa = 11.24SYY103 pKa = 10.79IDD105 pKa = 3.7KK106 pKa = 11.33DD107 pKa = 3.49GDD109 pKa = 3.65TLEE112 pKa = 4.14WGEE115 pKa = 3.84IQIDD119 pKa = 3.71GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 4.41AYY134 pKa = 9.38PPQRR138 pKa = 11.84FTQAVSQRR146 pKa = 11.84LLQYY150 pKa = 10.83LEE152 pKa = 4.55NYY154 pKa = 8.24PLRR157 pKa = 11.84IMFYY161 pKa = 10.62NFIIYY166 pKa = 8.54MLIYY170 pKa = 10.23IGFSHH175 pKa = 7.13LLWRR179 pKa = 11.84FIFLLFLLLLSIKK192 pKa = 10.35FPTNLKK198 pKa = 9.39SGRR201 pKa = 11.84VRR203 pKa = 11.84MSWRR207 pKa = 11.84PLRR210 pKa = 11.84GLGGPSVLWLKK221 pKa = 9.1VTVEE225 pKa = 3.97RR226 pKa = 11.84GRR228 pKa = 11.84RR229 pKa = 11.84CGPGHH234 pKa = 6.43WVHH237 pKa = 7.9IIISVGIWTSAQRR250 pKa = 11.84YY251 pKa = 5.06TVTTLGTTSLMTLIPIISNTLKK273 pKa = 10.86SLWGPRR279 pKa = 11.84EE280 pKa = 3.81TGKK283 pKa = 10.62AIPSTASQFKK293 pKa = 10.44LKK295 pKa = 10.77AGFQQSSFAIQGPIPAIKK313 pKa = 10.58NSWTRR318 pKa = 11.84KK319 pKa = 9.19RR320 pKa = 11.84IPLSRR325 pKa = 11.84PGHH328 pKa = 5.5FKK330 pKa = 10.77CVVRR334 pKa = 11.84YY335 pKa = 7.95PRR337 pKa = 11.84RR338 pKa = 11.84GSLLRR343 pKa = 11.84CPSKK347 pKa = 10.69SSIRR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84RR354 pKa = 11.84GKK356 pKa = 10.06YY357 pKa = 9.64AGRR360 pKa = 11.84GG361 pKa = 3.46
Molecular weight: 41.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1167 |
98 |
361 |
194.5 |
22.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.027 ± 0.436 | 2.399 ± 0.216 |
3.085 ± 0.598 | 3.77 ± 0.736 |
4.456 ± 0.509 | 5.998 ± 0.625 |
3.171 ± 0.781 | 5.827 ± 0.924 |
4.542 ± 0.967 | 8.74 ± 1.007 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.742 ± 0.724 | 3.599 ± 0.425 |
5.656 ± 0.391 | 5.056 ± 0.677 |
9.169 ± 0.936 | 9.854 ± 1.366 |
7.626 ± 1.072 | 5.398 ± 1.095 |
1.885 ± 0.252 | 2.999 ± 0.498 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
