
Roseburia sp. CAG:45
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; environmental samples
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
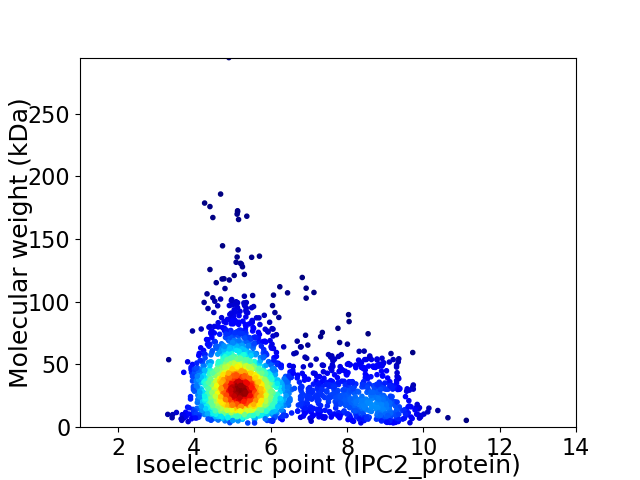
Virtual 2D-PAGE plot for 2315 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6NDA0|R6NDA0_9FIRM Transcriptional regulator MarR family OS=Roseburia sp. CAG:45 OX=1262947 GN=BN662_01280 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.26KK3 pKa = 10.38KK4 pKa = 10.72LISTLLCVSMVAAMVAGCGSGDD26 pKa = 3.66DD27 pKa = 4.1TKK29 pKa = 11.36DD30 pKa = 3.51ANNDD34 pKa = 3.09ASNTEE39 pKa = 4.03GTADD43 pKa = 4.34ADD45 pKa = 3.83TDD47 pKa = 3.82TDD49 pKa = 3.8SGSDD53 pKa = 3.33STEE56 pKa = 3.75VEE58 pKa = 3.96NTITGDD64 pKa = 3.57PSADD68 pKa = 3.36DD69 pKa = 4.85AFVVWGWNDD78 pKa = 3.8DD79 pKa = 3.35IKK81 pKa = 11.45NILDD85 pKa = 3.66GPFAEE90 pKa = 5.14QYY92 pKa = 10.09PDD94 pKa = 3.12LAKK97 pKa = 10.57RR98 pKa = 11.84IVFVNAGGSDD108 pKa = 4.07YY109 pKa = 10.9YY110 pKa = 8.83QTKK113 pKa = 9.63IDD115 pKa = 5.09EE116 pKa = 4.66ILDD119 pKa = 4.14DD120 pKa = 5.29PDD122 pKa = 4.42NEE124 pKa = 4.8LYY126 pKa = 9.9PDD128 pKa = 4.09LMGLEE133 pKa = 3.98VDD135 pKa = 4.07YY136 pKa = 11.22VLKK139 pKa = 10.78YY140 pKa = 11.07VNSDD144 pKa = 2.6WLTSVGDD151 pKa = 4.07CGITADD157 pKa = 5.17DD158 pKa = 4.17YY159 pKa = 12.05ANQYY163 pKa = 9.89QYY165 pKa = 11.82NLDD168 pKa = 3.83LGTVSGEE175 pKa = 4.28DD176 pKa = 3.12SSDD179 pKa = 3.56ANNVKK184 pKa = 10.48ALFWQATPGCFQLRR198 pKa = 11.84ADD200 pKa = 4.07LCEE203 pKa = 4.77KK204 pKa = 10.89YY205 pKa = 10.79LGTTDD210 pKa = 3.76PAEE213 pKa = 4.31LSTMFSTWDD222 pKa = 4.15GILEE226 pKa = 4.25AARR229 pKa = 11.84KK230 pKa = 10.03VNDD233 pKa = 3.89ASSGKK238 pKa = 10.12CKK240 pKa = 10.37LFSGYY245 pKa = 9.58DD246 pKa = 2.98EE247 pKa = 4.7CFRR250 pKa = 11.84VLSNSRR256 pKa = 11.84ATGWYY261 pKa = 10.02GDD263 pKa = 4.36DD264 pKa = 5.14DD265 pKa = 5.61VITVDD270 pKa = 5.47DD271 pKa = 4.52NMTEE275 pKa = 4.05YY276 pKa = 10.42MDD278 pKa = 4.17LAKK281 pKa = 10.42TMVDD285 pKa = 2.88EE286 pKa = 4.56GLTYY290 pKa = 9.62EE291 pKa = 4.13TDD293 pKa = 2.78QWSTDD298 pKa = 2.44WYY300 pKa = 10.94ANMEE304 pKa = 4.31GDD306 pKa = 4.65GEE308 pKa = 4.45TSNAAVAYY316 pKa = 8.55CGCPWFTYY324 pKa = 9.57WSLKK328 pKa = 9.1DD329 pKa = 3.13TWKK332 pKa = 11.22GNTILVNAPEE342 pKa = 3.88QFYY345 pKa = 10.47WGGTGLAATANCADD359 pKa = 4.14KK360 pKa = 11.13EE361 pKa = 4.26MAGTIIKK368 pKa = 10.6AFTCDD373 pKa = 2.84TDD375 pKa = 4.49FMVSINAKK383 pKa = 10.42NSDD386 pKa = 3.39FVNNKK391 pKa = 9.03EE392 pKa = 4.02AVKK395 pKa = 10.39KK396 pKa = 10.39ISEE399 pKa = 4.28AGATCDD405 pKa = 3.72YY406 pKa = 10.32LYY408 pKa = 10.82EE409 pKa = 4.43AAGQDD414 pKa = 2.66IMAFYY419 pKa = 11.04LPMADD424 pKa = 5.11SISAKK429 pKa = 10.3NATAEE434 pKa = 4.06DD435 pKa = 3.92QNINSAWSTQVKK447 pKa = 9.92EE448 pKa = 3.98YY449 pKa = 11.05AAGNKK454 pKa = 10.24DD455 pKa = 3.71KK456 pKa = 10.39DD457 pKa = 3.54TAIADD462 pKa = 4.19FKK464 pKa = 11.35SAVHH468 pKa = 7.06DD469 pKa = 3.94SYY471 pKa = 12.12SYY473 pKa = 11.45LKK475 pKa = 11.11AEE477 pKa = 4.2
MM1 pKa = 7.45KK2 pKa = 10.26KK3 pKa = 10.38KK4 pKa = 10.72LISTLLCVSMVAAMVAGCGSGDD26 pKa = 3.66DD27 pKa = 4.1TKK29 pKa = 11.36DD30 pKa = 3.51ANNDD34 pKa = 3.09ASNTEE39 pKa = 4.03GTADD43 pKa = 4.34ADD45 pKa = 3.83TDD47 pKa = 3.82TDD49 pKa = 3.8SGSDD53 pKa = 3.33STEE56 pKa = 3.75VEE58 pKa = 3.96NTITGDD64 pKa = 3.57PSADD68 pKa = 3.36DD69 pKa = 4.85AFVVWGWNDD78 pKa = 3.8DD79 pKa = 3.35IKK81 pKa = 11.45NILDD85 pKa = 3.66GPFAEE90 pKa = 5.14QYY92 pKa = 10.09PDD94 pKa = 3.12LAKK97 pKa = 10.57RR98 pKa = 11.84IVFVNAGGSDD108 pKa = 4.07YY109 pKa = 10.9YY110 pKa = 8.83QTKK113 pKa = 9.63IDD115 pKa = 5.09EE116 pKa = 4.66ILDD119 pKa = 4.14DD120 pKa = 5.29PDD122 pKa = 4.42NEE124 pKa = 4.8LYY126 pKa = 9.9PDD128 pKa = 4.09LMGLEE133 pKa = 3.98VDD135 pKa = 4.07YY136 pKa = 11.22VLKK139 pKa = 10.78YY140 pKa = 11.07VNSDD144 pKa = 2.6WLTSVGDD151 pKa = 4.07CGITADD157 pKa = 5.17DD158 pKa = 4.17YY159 pKa = 12.05ANQYY163 pKa = 9.89QYY165 pKa = 11.82NLDD168 pKa = 3.83LGTVSGEE175 pKa = 4.28DD176 pKa = 3.12SSDD179 pKa = 3.56ANNVKK184 pKa = 10.48ALFWQATPGCFQLRR198 pKa = 11.84ADD200 pKa = 4.07LCEE203 pKa = 4.77KK204 pKa = 10.89YY205 pKa = 10.79LGTTDD210 pKa = 3.76PAEE213 pKa = 4.31LSTMFSTWDD222 pKa = 4.15GILEE226 pKa = 4.25AARR229 pKa = 11.84KK230 pKa = 10.03VNDD233 pKa = 3.89ASSGKK238 pKa = 10.12CKK240 pKa = 10.37LFSGYY245 pKa = 9.58DD246 pKa = 2.98EE247 pKa = 4.7CFRR250 pKa = 11.84VLSNSRR256 pKa = 11.84ATGWYY261 pKa = 10.02GDD263 pKa = 4.36DD264 pKa = 5.14DD265 pKa = 5.61VITVDD270 pKa = 5.47DD271 pKa = 4.52NMTEE275 pKa = 4.05YY276 pKa = 10.42MDD278 pKa = 4.17LAKK281 pKa = 10.42TMVDD285 pKa = 2.88EE286 pKa = 4.56GLTYY290 pKa = 9.62EE291 pKa = 4.13TDD293 pKa = 2.78QWSTDD298 pKa = 2.44WYY300 pKa = 10.94ANMEE304 pKa = 4.31GDD306 pKa = 4.65GEE308 pKa = 4.45TSNAAVAYY316 pKa = 8.55CGCPWFTYY324 pKa = 9.57WSLKK328 pKa = 9.1DD329 pKa = 3.13TWKK332 pKa = 11.22GNTILVNAPEE342 pKa = 3.88QFYY345 pKa = 10.47WGGTGLAATANCADD359 pKa = 4.14KK360 pKa = 11.13EE361 pKa = 4.26MAGTIIKK368 pKa = 10.6AFTCDD373 pKa = 2.84TDD375 pKa = 4.49FMVSINAKK383 pKa = 10.42NSDD386 pKa = 3.39FVNNKK391 pKa = 9.03EE392 pKa = 4.02AVKK395 pKa = 10.39KK396 pKa = 10.39ISEE399 pKa = 4.28AGATCDD405 pKa = 3.72YY406 pKa = 10.32LYY408 pKa = 10.82EE409 pKa = 4.43AAGQDD414 pKa = 2.66IMAFYY419 pKa = 11.04LPMADD424 pKa = 5.11SISAKK429 pKa = 10.3NATAEE434 pKa = 4.06DD435 pKa = 3.92QNINSAWSTQVKK447 pKa = 9.92EE448 pKa = 3.98YY449 pKa = 11.05AAGNKK454 pKa = 10.24DD455 pKa = 3.71KK456 pKa = 10.39DD457 pKa = 3.54TAIADD462 pKa = 4.19FKK464 pKa = 11.35SAVHH468 pKa = 7.06DD469 pKa = 3.94SYY471 pKa = 12.12SYY473 pKa = 11.45LKK475 pKa = 11.11AEE477 pKa = 4.2
Molecular weight: 52.12 kDa
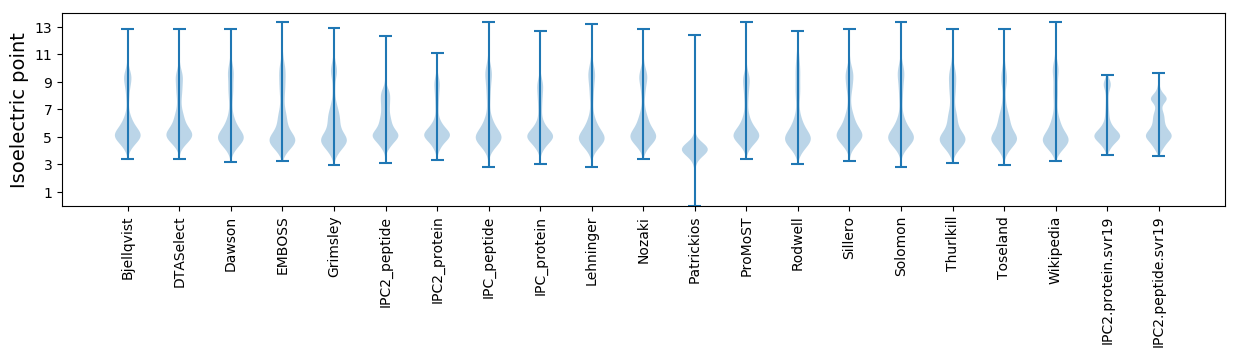
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6NZ56|R6NZ56_9FIRM Chorismate synthase OS=Roseburia sp. CAG:45 OX=1262947 GN=aroC PE=3 SV=1
MM1 pKa = 7.64HH2 pKa = 7.44KK3 pKa = 10.88GKK5 pKa = 8.15MTFQPKK11 pKa = 8.98NRR13 pKa = 11.84QRR15 pKa = 11.84NKK17 pKa = 7.97VHH19 pKa = 6.65GFRR22 pKa = 11.84KK23 pKa = 9.94RR24 pKa = 11.84MSTAGGRR31 pKa = 11.84KK32 pKa = 8.62VLAARR37 pKa = 11.84RR38 pKa = 11.84LKK40 pKa = 10.61GRR42 pKa = 11.84KK43 pKa = 8.87KK44 pKa = 10.59LSAA47 pKa = 3.95
MM1 pKa = 7.64HH2 pKa = 7.44KK3 pKa = 10.88GKK5 pKa = 8.15MTFQPKK11 pKa = 8.98NRR13 pKa = 11.84QRR15 pKa = 11.84NKK17 pKa = 7.97VHH19 pKa = 6.65GFRR22 pKa = 11.84KK23 pKa = 9.94RR24 pKa = 11.84MSTAGGRR31 pKa = 11.84KK32 pKa = 8.62VLAARR37 pKa = 11.84RR38 pKa = 11.84LKK40 pKa = 10.61GRR42 pKa = 11.84KK43 pKa = 8.87KK44 pKa = 10.59LSAA47 pKa = 3.95
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
734199 |
29 |
2707 |
317.1 |
35.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.622 ± 0.049 | 1.404 ± 0.021 |
5.682 ± 0.047 | 8.047 ± 0.057 |
3.972 ± 0.042 | 6.967 ± 0.046 |
1.768 ± 0.025 | 7.54 ± 0.052 |
7.034 ± 0.045 | 8.824 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.201 ± 0.028 | 4.481 ± 0.038 |
3.062 ± 0.029 | 3.257 ± 0.032 |
4.061 ± 0.038 | 5.623 ± 0.046 |
5.48 ± 0.041 | 6.919 ± 0.045 |
0.789 ± 0.018 | 4.266 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
