
Cyanophage S-RIM14
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Ahtivirus; unclassified Ahtivirus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
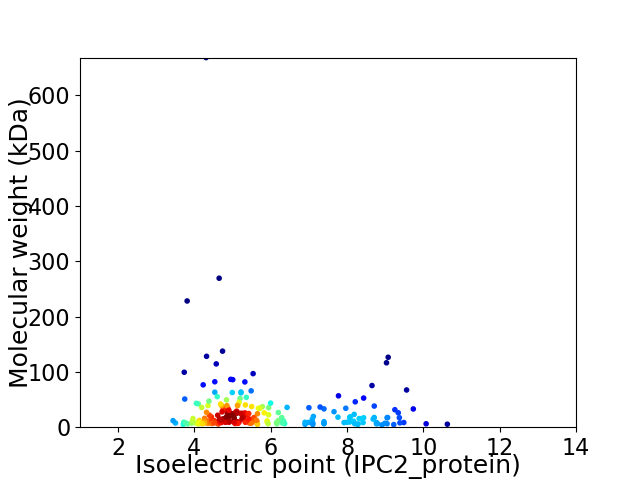
Virtual 2D-PAGE plot for 216 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D7SJR7|A0A1D7SJR7_9CAUD Uncharacterized protein OS=Cyanophage S-RIM14 OX=1278423 GN=LIS021110_020 PE=4 SV=1
MM1 pKa = 7.26TLYY4 pKa = 10.52RR5 pKa = 11.84PNGDD9 pKa = 3.97FGPVCDD15 pKa = 4.74PFEE18 pKa = 4.47EE19 pKa = 4.57PTDD22 pKa = 3.85FSLPILADD30 pKa = 3.71PLVLAFPSPQRR41 pKa = 11.84VLVDD45 pKa = 4.17LEE47 pKa = 4.19GQTVVNVPVWNDD59 pKa = 3.3PGDD62 pKa = 3.74YY63 pKa = 10.19RR64 pKa = 11.84PNGSMGPVCDD74 pKa = 3.66PTSPYY79 pKa = 10.56GPEE82 pKa = 3.9DD83 pKa = 3.78SIGEE87 pKa = 4.03SDD89 pKa = 4.07PRR91 pKa = 11.84VVDD94 pKa = 3.9PQAPPEE100 pKa = 4.57GGGPAYY106 pKa = 10.54SGPTEE111 pKa = 4.28PYY113 pKa = 10.15RR114 pKa = 11.84PLGDD118 pKa = 3.77MGPVCDD124 pKa = 4.57PFVPVNLNPEE134 pKa = 4.34VVTPRR139 pKa = 11.84QDD141 pKa = 3.63LDD143 pKa = 3.85GPAADD148 pKa = 5.31DD149 pKa = 5.34DD150 pKa = 5.14DD151 pKa = 6.71GEE153 pKa = 4.74TYY155 pKa = 10.43PDD157 pKa = 4.65PRR159 pKa = 11.84VQDD162 pKa = 3.72PKK164 pKa = 10.56RR165 pKa = 11.84PSSVPVSIVEE175 pKa = 4.17PPDD178 pKa = 3.38YY179 pKa = 10.93RR180 pKa = 11.84PDD182 pKa = 3.67GPFGPVCDD190 pKa = 4.15PTLDD194 pKa = 3.97PLLTATAGDD203 pKa = 3.79NAIGDD208 pKa = 4.27GNPDD212 pKa = 4.39DD213 pKa = 5.8LNPDD217 pKa = 4.43DD218 pKa = 5.22IYY220 pKa = 11.07PDD222 pKa = 3.24IVNFPAALPNWTNLPDD238 pKa = 3.87SPPTIGLPSTDD249 pKa = 3.68EE250 pKa = 4.3FEE252 pKa = 5.8DD253 pKa = 5.61GIDD256 pKa = 3.69APSVAGVAVSYY267 pKa = 10.93IPFFDD272 pKa = 3.41TSNINNIPASFSFDD286 pKa = 3.8DD287 pKa = 4.29PEE289 pKa = 5.41PDD291 pKa = 3.3KK292 pKa = 11.72GDD294 pKa = 3.96WNWSHH299 pKa = 6.91YY300 pKa = 9.61YY301 pKa = 10.67FNGKK305 pKa = 8.78FFEE308 pKa = 4.27WDD310 pKa = 3.08EE311 pKa = 4.14TDD313 pKa = 3.22TFNGLTGAGLYY324 pKa = 10.9GEE326 pKa = 5.78LIDD329 pKa = 5.66AADD332 pKa = 4.17NDD334 pKa = 4.14VEE336 pKa = 4.73IPDD339 pKa = 3.62GDD341 pKa = 3.58RR342 pKa = 11.84EE343 pKa = 4.35VFQPDD348 pKa = 4.25SYY350 pKa = 11.63CEE352 pKa = 4.13EE353 pKa = 3.97PWGDD357 pKa = 3.72QVLDD361 pKa = 4.64PYY363 pKa = 11.16TDD365 pKa = 3.19TDD367 pKa = 3.82GNVVYY372 pKa = 10.33RR373 pKa = 11.84QVRR376 pKa = 11.84SFPRR380 pKa = 11.84TYY382 pKa = 10.22QVSHH386 pKa = 6.57EE387 pKa = 4.13NFPEE391 pKa = 3.93EE392 pKa = 4.24FAQKK396 pKa = 10.22SAWISQGICSFEE408 pKa = 3.98GKK410 pKa = 7.83TRR412 pKa = 11.84EE413 pKa = 4.46AGDD416 pKa = 3.25AAYY419 pKa = 7.42FTSTITIPDD428 pKa = 3.34TDD430 pKa = 3.9TYY432 pKa = 10.5KK433 pKa = 10.28IRR435 pKa = 11.84YY436 pKa = 8.7KK437 pKa = 10.27FFKK440 pKa = 10.22RR441 pKa = 11.84GEE443 pKa = 3.89IFLDD447 pKa = 3.56YY448 pKa = 10.71FDD450 pKa = 5.3PNKK453 pKa = 8.46TTIEE457 pKa = 4.12LVSEE461 pKa = 4.52AEE463 pKa = 4.42GSASFSGDD471 pKa = 2.94EE472 pKa = 3.96YY473 pKa = 11.24DD474 pKa = 3.73VVSRR478 pKa = 11.84EE479 pKa = 4.05IEE481 pKa = 3.94AGEE484 pKa = 3.91YY485 pKa = 10.88DD486 pKa = 3.74LTTFIEE492 pKa = 4.7NYY494 pKa = 10.64AIGTPQSLWKK504 pKa = 10.41DD505 pKa = 3.59SPAACAIEE513 pKa = 5.13VYY515 pKa = 10.48QGDD518 pKa = 3.95FSIVTGQIPCDD529 pKa = 3.48VGIIEE534 pKa = 4.52SNIPGGASFDD544 pKa = 4.29SNGNIIVTGQCRR556 pKa = 11.84VQFDD560 pKa = 3.75FSYY563 pKa = 11.42DD564 pKa = 4.0DD565 pKa = 4.49NPDD568 pKa = 3.32TYY570 pKa = 10.28GTALGTVSYY579 pKa = 10.43SALNISFTQDD589 pKa = 2.84TQTSTGSARR598 pKa = 11.84EE599 pKa = 4.05TRR601 pKa = 11.84RR602 pKa = 11.84VAQGTYY608 pKa = 8.95QVSLSNANSAGFVLQNNNTEE628 pKa = 3.68ICFFDD633 pKa = 5.95GDD635 pKa = 4.1AQDD638 pKa = 4.61CNALLRR644 pKa = 11.84LTTLDD649 pKa = 3.39STSAGEE655 pKa = 4.88GVTFTCGTVAEE666 pKa = 4.51VEE668 pKa = 3.92LGLRR672 pKa = 11.84YY673 pKa = 10.08DD674 pKa = 5.3DD675 pKa = 5.08NPTTQGLCLSSVTWSGDD692 pKa = 2.91EE693 pKa = 5.26DD694 pKa = 3.48ITLVRR699 pKa = 11.84DD700 pKa = 3.61TNKK703 pKa = 10.22EE704 pKa = 3.97SGSRR708 pKa = 11.84TVRR711 pKa = 11.84KK712 pKa = 9.85DD713 pKa = 3.55LLSGTYY719 pKa = 9.6TMNLVDD725 pKa = 3.76NAGGYY730 pKa = 9.44LVQNPGTTQNDD741 pKa = 4.27SYY743 pKa = 11.42IVLYY747 pKa = 10.83DD748 pKa = 3.67GGGSDD753 pKa = 3.47EE754 pKa = 4.52NGRR757 pKa = 11.84LTCEE761 pKa = 4.18QISATGSSVFSSFDD775 pKa = 3.57SNGNLVITGTRR786 pKa = 11.84GYY788 pKa = 9.29TNDD791 pKa = 3.96FIEE794 pKa = 4.57GFKK797 pKa = 10.7DD798 pKa = 4.91DD799 pKa = 3.81SSVWQQSDD807 pKa = 3.45VGTEE811 pKa = 3.69ILFNATDD818 pKa = 3.58SSKK821 pKa = 11.1GMQVQIGIIPRR832 pKa = 11.84NRR834 pKa = 11.84YY835 pKa = 9.24DD836 pKa = 3.43GNNYY840 pKa = 8.8TISTDD845 pKa = 3.08WAVKK849 pKa = 9.87QIINYY854 pKa = 8.66GVGYY858 pKa = 10.43SVDD861 pKa = 3.77DD862 pKa = 3.68TWNVSYY868 pKa = 10.77TNSATGGTTSAKK880 pKa = 10.35VKK882 pKa = 10.27IFSTRR887 pKa = 11.84KK888 pKa = 9.26GRR890 pKa = 11.84ATVGNLVWSTTNNAVGYY907 pKa = 9.52DD908 pKa = 3.58EE909 pKa = 6.03SFTPFPP915 pKa = 4.71
MM1 pKa = 7.26TLYY4 pKa = 10.52RR5 pKa = 11.84PNGDD9 pKa = 3.97FGPVCDD15 pKa = 4.74PFEE18 pKa = 4.47EE19 pKa = 4.57PTDD22 pKa = 3.85FSLPILADD30 pKa = 3.71PLVLAFPSPQRR41 pKa = 11.84VLVDD45 pKa = 4.17LEE47 pKa = 4.19GQTVVNVPVWNDD59 pKa = 3.3PGDD62 pKa = 3.74YY63 pKa = 10.19RR64 pKa = 11.84PNGSMGPVCDD74 pKa = 3.66PTSPYY79 pKa = 10.56GPEE82 pKa = 3.9DD83 pKa = 3.78SIGEE87 pKa = 4.03SDD89 pKa = 4.07PRR91 pKa = 11.84VVDD94 pKa = 3.9PQAPPEE100 pKa = 4.57GGGPAYY106 pKa = 10.54SGPTEE111 pKa = 4.28PYY113 pKa = 10.15RR114 pKa = 11.84PLGDD118 pKa = 3.77MGPVCDD124 pKa = 4.57PFVPVNLNPEE134 pKa = 4.34VVTPRR139 pKa = 11.84QDD141 pKa = 3.63LDD143 pKa = 3.85GPAADD148 pKa = 5.31DD149 pKa = 5.34DD150 pKa = 5.14DD151 pKa = 6.71GEE153 pKa = 4.74TYY155 pKa = 10.43PDD157 pKa = 4.65PRR159 pKa = 11.84VQDD162 pKa = 3.72PKK164 pKa = 10.56RR165 pKa = 11.84PSSVPVSIVEE175 pKa = 4.17PPDD178 pKa = 3.38YY179 pKa = 10.93RR180 pKa = 11.84PDD182 pKa = 3.67GPFGPVCDD190 pKa = 4.15PTLDD194 pKa = 3.97PLLTATAGDD203 pKa = 3.79NAIGDD208 pKa = 4.27GNPDD212 pKa = 4.39DD213 pKa = 5.8LNPDD217 pKa = 4.43DD218 pKa = 5.22IYY220 pKa = 11.07PDD222 pKa = 3.24IVNFPAALPNWTNLPDD238 pKa = 3.87SPPTIGLPSTDD249 pKa = 3.68EE250 pKa = 4.3FEE252 pKa = 5.8DD253 pKa = 5.61GIDD256 pKa = 3.69APSVAGVAVSYY267 pKa = 10.93IPFFDD272 pKa = 3.41TSNINNIPASFSFDD286 pKa = 3.8DD287 pKa = 4.29PEE289 pKa = 5.41PDD291 pKa = 3.3KK292 pKa = 11.72GDD294 pKa = 3.96WNWSHH299 pKa = 6.91YY300 pKa = 9.61YY301 pKa = 10.67FNGKK305 pKa = 8.78FFEE308 pKa = 4.27WDD310 pKa = 3.08EE311 pKa = 4.14TDD313 pKa = 3.22TFNGLTGAGLYY324 pKa = 10.9GEE326 pKa = 5.78LIDD329 pKa = 5.66AADD332 pKa = 4.17NDD334 pKa = 4.14VEE336 pKa = 4.73IPDD339 pKa = 3.62GDD341 pKa = 3.58RR342 pKa = 11.84EE343 pKa = 4.35VFQPDD348 pKa = 4.25SYY350 pKa = 11.63CEE352 pKa = 4.13EE353 pKa = 3.97PWGDD357 pKa = 3.72QVLDD361 pKa = 4.64PYY363 pKa = 11.16TDD365 pKa = 3.19TDD367 pKa = 3.82GNVVYY372 pKa = 10.33RR373 pKa = 11.84QVRR376 pKa = 11.84SFPRR380 pKa = 11.84TYY382 pKa = 10.22QVSHH386 pKa = 6.57EE387 pKa = 4.13NFPEE391 pKa = 3.93EE392 pKa = 4.24FAQKK396 pKa = 10.22SAWISQGICSFEE408 pKa = 3.98GKK410 pKa = 7.83TRR412 pKa = 11.84EE413 pKa = 4.46AGDD416 pKa = 3.25AAYY419 pKa = 7.42FTSTITIPDD428 pKa = 3.34TDD430 pKa = 3.9TYY432 pKa = 10.5KK433 pKa = 10.28IRR435 pKa = 11.84YY436 pKa = 8.7KK437 pKa = 10.27FFKK440 pKa = 10.22RR441 pKa = 11.84GEE443 pKa = 3.89IFLDD447 pKa = 3.56YY448 pKa = 10.71FDD450 pKa = 5.3PNKK453 pKa = 8.46TTIEE457 pKa = 4.12LVSEE461 pKa = 4.52AEE463 pKa = 4.42GSASFSGDD471 pKa = 2.94EE472 pKa = 3.96YY473 pKa = 11.24DD474 pKa = 3.73VVSRR478 pKa = 11.84EE479 pKa = 4.05IEE481 pKa = 3.94AGEE484 pKa = 3.91YY485 pKa = 10.88DD486 pKa = 3.74LTTFIEE492 pKa = 4.7NYY494 pKa = 10.64AIGTPQSLWKK504 pKa = 10.41DD505 pKa = 3.59SPAACAIEE513 pKa = 5.13VYY515 pKa = 10.48QGDD518 pKa = 3.95FSIVTGQIPCDD529 pKa = 3.48VGIIEE534 pKa = 4.52SNIPGGASFDD544 pKa = 4.29SNGNIIVTGQCRR556 pKa = 11.84VQFDD560 pKa = 3.75FSYY563 pKa = 11.42DD564 pKa = 4.0DD565 pKa = 4.49NPDD568 pKa = 3.32TYY570 pKa = 10.28GTALGTVSYY579 pKa = 10.43SALNISFTQDD589 pKa = 2.84TQTSTGSARR598 pKa = 11.84EE599 pKa = 4.05TRR601 pKa = 11.84RR602 pKa = 11.84VAQGTYY608 pKa = 8.95QVSLSNANSAGFVLQNNNTEE628 pKa = 3.68ICFFDD633 pKa = 5.95GDD635 pKa = 4.1AQDD638 pKa = 4.61CNALLRR644 pKa = 11.84LTTLDD649 pKa = 3.39STSAGEE655 pKa = 4.88GVTFTCGTVAEE666 pKa = 4.51VEE668 pKa = 3.92LGLRR672 pKa = 11.84YY673 pKa = 10.08DD674 pKa = 5.3DD675 pKa = 5.08NPTTQGLCLSSVTWSGDD692 pKa = 2.91EE693 pKa = 5.26DD694 pKa = 3.48ITLVRR699 pKa = 11.84DD700 pKa = 3.61TNKK703 pKa = 10.22EE704 pKa = 3.97SGSRR708 pKa = 11.84TVRR711 pKa = 11.84KK712 pKa = 9.85DD713 pKa = 3.55LLSGTYY719 pKa = 9.6TMNLVDD725 pKa = 3.76NAGGYY730 pKa = 9.44LVQNPGTTQNDD741 pKa = 4.27SYY743 pKa = 11.42IVLYY747 pKa = 10.83DD748 pKa = 3.67GGGSDD753 pKa = 3.47EE754 pKa = 4.52NGRR757 pKa = 11.84LTCEE761 pKa = 4.18QISATGSSVFSSFDD775 pKa = 3.57SNGNLVITGTRR786 pKa = 11.84GYY788 pKa = 9.29TNDD791 pKa = 3.96FIEE794 pKa = 4.57GFKK797 pKa = 10.7DD798 pKa = 4.91DD799 pKa = 3.81SSVWQQSDD807 pKa = 3.45VGTEE811 pKa = 3.69ILFNATDD818 pKa = 3.58SSKK821 pKa = 11.1GMQVQIGIIPRR832 pKa = 11.84NRR834 pKa = 11.84YY835 pKa = 9.24DD836 pKa = 3.43GNNYY840 pKa = 8.8TISTDD845 pKa = 3.08WAVKK849 pKa = 9.87QIINYY854 pKa = 8.66GVGYY858 pKa = 10.43SVDD861 pKa = 3.77DD862 pKa = 3.68TWNVSYY868 pKa = 10.77TNSATGGTTSAKK880 pKa = 10.35VKK882 pKa = 10.27IFSTRR887 pKa = 11.84KK888 pKa = 9.26GRR890 pKa = 11.84ATVGNLVWSTTNNAVGYY907 pKa = 9.52DD908 pKa = 3.58EE909 pKa = 6.03SFTPFPP915 pKa = 4.71
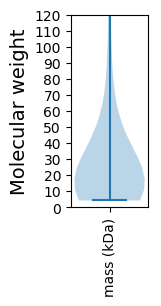
Molecular weight: 99.35 kDa
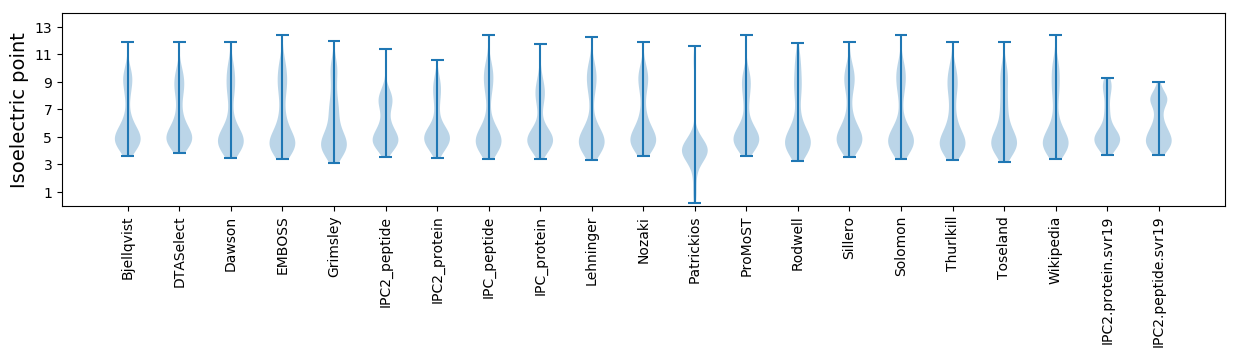
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D7SHV3|A0A1D7SHV3_9CAUD Uncharacterized protein OS=Cyanophage S-RIM14 OX=1278423 GN=LIS021110_046 PE=4 SV=1
MM1 pKa = 7.01TRR3 pKa = 11.84VRR5 pKa = 11.84NQVGTWFGTKK15 pKa = 7.43TTLDD19 pKa = 3.88DD20 pKa = 4.16KK21 pKa = 11.76SLGGLSANQKK31 pKa = 10.17RR32 pKa = 11.84EE33 pKa = 3.73VLRR36 pKa = 11.84LIDD39 pKa = 4.26RR40 pKa = 11.84AIDD43 pKa = 3.35NHH45 pKa = 5.87NRR47 pKa = 11.84TATLVSASIGGVLLFFYY64 pKa = 10.68AHH66 pKa = 6.07GVVSIVDD73 pKa = 3.65RR74 pKa = 11.84TANN77 pKa = 3.21
MM1 pKa = 7.01TRR3 pKa = 11.84VRR5 pKa = 11.84NQVGTWFGTKK15 pKa = 7.43TTLDD19 pKa = 3.88DD20 pKa = 4.16KK21 pKa = 11.76SLGGLSANQKK31 pKa = 10.17RR32 pKa = 11.84EE33 pKa = 3.73VLRR36 pKa = 11.84LIDD39 pKa = 4.26RR40 pKa = 11.84AIDD43 pKa = 3.35NHH45 pKa = 5.87NRR47 pKa = 11.84TATLVSASIGGVLLFFYY64 pKa = 10.68AHH66 pKa = 6.07GVVSIVDD73 pKa = 3.65RR74 pKa = 11.84TANN77 pKa = 3.21
Molecular weight: 8.44 kDa
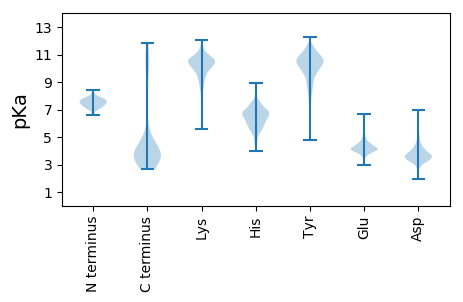
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
57942 |
34 |
6198 |
268.3 |
29.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.902 ± 0.246 | 0.951 ± 0.1 |
6.765 ± 0.151 | 6.047 ± 0.29 |
4.28 ± 0.1 | 7.956 ± 0.37 |
1.436 ± 0.123 | 6.325 ± 0.198 |
5.645 ± 0.388 | 7.042 ± 0.172 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.188 ± 0.194 | 5.726 ± 0.157 |
4.025 ± 0.15 | 3.862 ± 0.097 |
4.237 ± 0.198 | 7.159 ± 0.192 |
7.397 ± 0.462 | 6.615 ± 0.168 |
1.179 ± 0.093 | 4.261 ± 0.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
