
Changjiang tombus-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.8
Get precalculated fractions of proteins
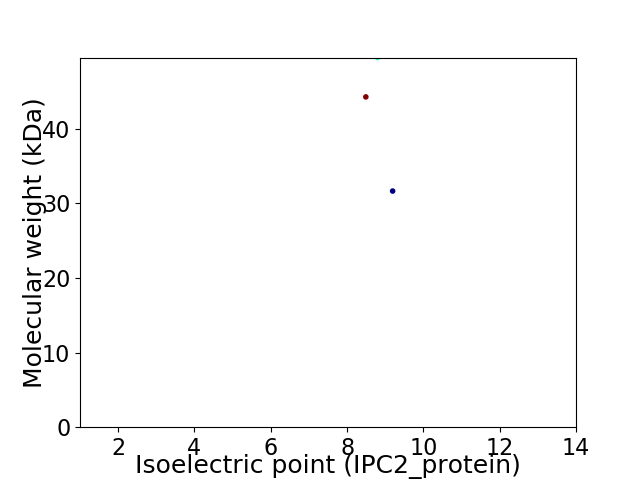
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG06|A0A1L3KG06_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 18 OX=1922811 PE=4 SV=1
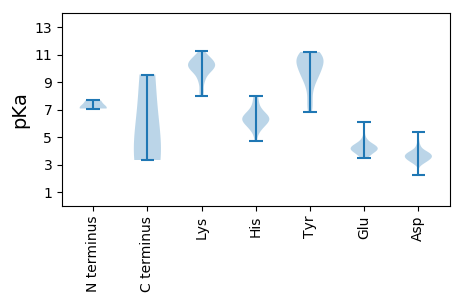
MM1 pKa = 7.66FKK3 pKa = 9.91TLNQKK8 pKa = 9.25QLALARR14 pKa = 11.84TRR16 pKa = 11.84SKK18 pKa = 10.85KK19 pKa = 10.02ILANRR24 pKa = 11.84YY25 pKa = 8.44LNCRR29 pKa = 11.84YY30 pKa = 10.37NPFEE34 pKa = 4.16SRR36 pKa = 11.84GGAMIPDD43 pKa = 5.35GIGKK47 pKa = 8.97HH48 pKa = 5.52LEE50 pKa = 4.0SRR52 pKa = 11.84DD53 pKa = 3.46FRR55 pKa = 11.84SIYY58 pKa = 9.04HH59 pKa = 6.33LKK61 pKa = 8.94VTGQAEE67 pKa = 4.48VLILPAIPMVAAVISPEE84 pKa = 4.01GNTLEE89 pKa = 4.19VNGMSIINGSYY100 pKa = 10.6VGLCGSNLLKK110 pKa = 11.13YY111 pKa = 7.17NTPLGGTVVQGDD123 pKa = 4.47DD124 pKa = 3.42VTSARR129 pKa = 11.84IVTVGYY135 pKa = 10.4RR136 pKa = 11.84LTYY139 pKa = 9.55EE140 pKa = 4.6GKK142 pKa = 8.27ATEE145 pKa = 4.4ANGQLVANSLPLMLDD160 pKa = 3.59GSPMLNSGSYY170 pKa = 9.93VFSGGNPVTKK180 pKa = 10.47VAGSMTAINIDD191 pKa = 3.81MEE193 pKa = 4.73TNAHH197 pKa = 4.68VTTRR201 pKa = 11.84DD202 pKa = 3.25TKK204 pKa = 10.32IFRR207 pKa = 11.84PEE209 pKa = 3.59EE210 pKa = 4.03GVAGVLKK217 pKa = 10.62RR218 pKa = 11.84QVTVANHH225 pKa = 5.72AFKK228 pKa = 10.6PFWEE232 pKa = 4.35TGVYY236 pKa = 9.84PNNLNGLDD244 pKa = 3.86NVSKK248 pKa = 10.65AAWLQNPPAGLVDD261 pKa = 4.05NNSISYY267 pKa = 7.53PTVAVLDD274 pKa = 3.81HH275 pKa = 6.72ALGVEE280 pKa = 4.41SIVINNDD287 pKa = 2.23NTTPLLYY294 pKa = 10.19RR295 pKa = 11.84LEE297 pKa = 5.16IIHH300 pKa = 6.57CVQFQHH306 pKa = 6.36SPNWGLADD314 pKa = 3.79LTVDD318 pKa = 3.98APAVDD323 pKa = 3.99KK324 pKa = 11.12QALEE328 pKa = 4.02ADD330 pKa = 3.84DD331 pKa = 4.8RR332 pKa = 11.84LNSTVTHH339 pKa = 6.32AVDD342 pKa = 3.7ATSSTIPTRR351 pKa = 11.84YY352 pKa = 9.8SSTVGPATSSIFSDD366 pKa = 3.27VTMDD370 pKa = 3.48MSTTKK375 pKa = 8.27TTTRR379 pKa = 11.84SPYY382 pKa = 10.98ARR384 pKa = 11.84TTTVLQQPKK393 pKa = 10.18RR394 pKa = 11.84KK395 pKa = 8.32ATTKK399 pKa = 9.21ATPSPAKK406 pKa = 10.3KK407 pKa = 10.06AGKK410 pKa = 9.56KK411 pKa = 9.55
MM1 pKa = 7.66FKK3 pKa = 9.91TLNQKK8 pKa = 9.25QLALARR14 pKa = 11.84TRR16 pKa = 11.84SKK18 pKa = 10.85KK19 pKa = 10.02ILANRR24 pKa = 11.84YY25 pKa = 8.44LNCRR29 pKa = 11.84YY30 pKa = 10.37NPFEE34 pKa = 4.16SRR36 pKa = 11.84GGAMIPDD43 pKa = 5.35GIGKK47 pKa = 8.97HH48 pKa = 5.52LEE50 pKa = 4.0SRR52 pKa = 11.84DD53 pKa = 3.46FRR55 pKa = 11.84SIYY58 pKa = 9.04HH59 pKa = 6.33LKK61 pKa = 8.94VTGQAEE67 pKa = 4.48VLILPAIPMVAAVISPEE84 pKa = 4.01GNTLEE89 pKa = 4.19VNGMSIINGSYY100 pKa = 10.6VGLCGSNLLKK110 pKa = 11.13YY111 pKa = 7.17NTPLGGTVVQGDD123 pKa = 4.47DD124 pKa = 3.42VTSARR129 pKa = 11.84IVTVGYY135 pKa = 10.4RR136 pKa = 11.84LTYY139 pKa = 9.55EE140 pKa = 4.6GKK142 pKa = 8.27ATEE145 pKa = 4.4ANGQLVANSLPLMLDD160 pKa = 3.59GSPMLNSGSYY170 pKa = 9.93VFSGGNPVTKK180 pKa = 10.47VAGSMTAINIDD191 pKa = 3.81MEE193 pKa = 4.73TNAHH197 pKa = 4.68VTTRR201 pKa = 11.84DD202 pKa = 3.25TKK204 pKa = 10.32IFRR207 pKa = 11.84PEE209 pKa = 3.59EE210 pKa = 4.03GVAGVLKK217 pKa = 10.62RR218 pKa = 11.84QVTVANHH225 pKa = 5.72AFKK228 pKa = 10.6PFWEE232 pKa = 4.35TGVYY236 pKa = 9.84PNNLNGLDD244 pKa = 3.86NVSKK248 pKa = 10.65AAWLQNPPAGLVDD261 pKa = 4.05NNSISYY267 pKa = 7.53PTVAVLDD274 pKa = 3.81HH275 pKa = 6.72ALGVEE280 pKa = 4.41SIVINNDD287 pKa = 2.23NTTPLLYY294 pKa = 10.19RR295 pKa = 11.84LEE297 pKa = 5.16IIHH300 pKa = 6.57CVQFQHH306 pKa = 6.36SPNWGLADD314 pKa = 3.79LTVDD318 pKa = 3.98APAVDD323 pKa = 3.99KK324 pKa = 11.12QALEE328 pKa = 4.02ADD330 pKa = 3.84DD331 pKa = 4.8RR332 pKa = 11.84LNSTVTHH339 pKa = 6.32AVDD342 pKa = 3.7ATSSTIPTRR351 pKa = 11.84YY352 pKa = 9.8SSTVGPATSSIFSDD366 pKa = 3.27VTMDD370 pKa = 3.48MSTTKK375 pKa = 8.27TTTRR379 pKa = 11.84SPYY382 pKa = 10.98ARR384 pKa = 11.84TTTVLQQPKK393 pKa = 10.18RR394 pKa = 11.84KK395 pKa = 8.32ATTKK399 pKa = 9.21ATPSPAKK406 pKa = 10.3KK407 pKa = 10.06AGKK410 pKa = 9.56KK411 pKa = 9.55
Molecular weight: 44.24 kDa
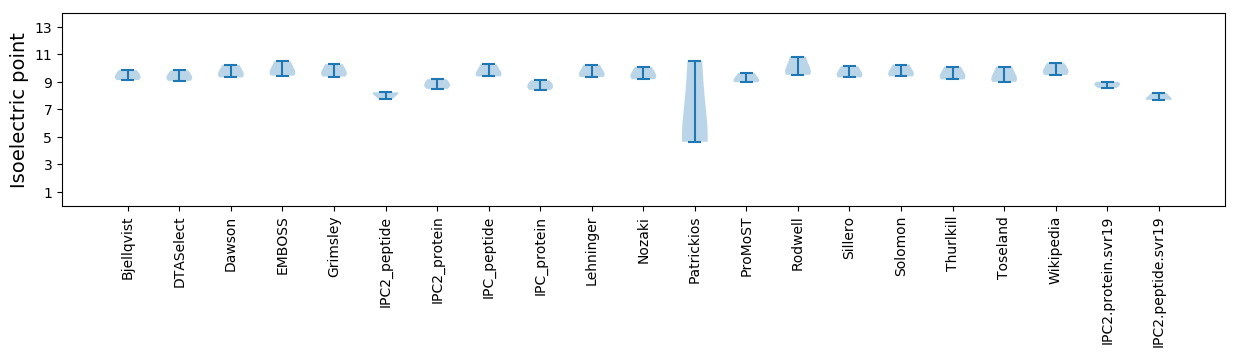
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFU2|A0A1L3KFU2_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 18 OX=1922811 PE=4 SV=1
MM1 pKa = 7.07MLSSSVNSPVTPALSAQTPQRR22 pKa = 11.84IGGIQSKK29 pKa = 7.97RR30 pKa = 11.84QKK32 pKa = 10.4VMGYY36 pKa = 9.68LRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.06LAKK43 pKa = 9.94VPYY46 pKa = 8.86HH47 pKa = 6.36VNPALTVRR55 pKa = 11.84DD56 pKa = 3.73LHH58 pKa = 8.02VDD60 pKa = 3.49EE61 pKa = 5.54PLSKK65 pKa = 10.24EE66 pKa = 3.44QTKK69 pKa = 10.09RR70 pKa = 11.84VEE72 pKa = 3.91EE73 pKa = 4.47FIEE76 pKa = 4.3VVEE79 pKa = 4.85DD80 pKa = 3.96PKK82 pKa = 11.27SSQALKK88 pKa = 10.41DD89 pKa = 3.26NRR91 pKa = 11.84YY92 pKa = 10.25AILEE96 pKa = 4.38DD97 pKa = 4.09EE98 pKa = 4.72VKK100 pKa = 9.33HH101 pKa = 6.18TIIQMPSPTGNTYY114 pKa = 9.45QVIRR118 pKa = 11.84NKK120 pKa = 8.82TPKK123 pKa = 9.27VKK125 pKa = 10.08KK126 pKa = 10.48VKK128 pKa = 9.7IDD130 pKa = 3.6SPVMPEE136 pKa = 3.65EE137 pKa = 4.18MFPDD141 pKa = 3.12RR142 pKa = 11.84QVINRR147 pKa = 11.84QVKK150 pKa = 10.2KK151 pKa = 10.75DD152 pKa = 3.68GILAKK157 pKa = 9.74TNPSLVYY164 pKa = 9.37YY165 pKa = 10.68LRR167 pKa = 11.84TKK169 pKa = 10.68FFLKK173 pKa = 10.56FRR175 pKa = 11.84DD176 pKa = 3.6HH177 pKa = 7.85ALMQTMVHH185 pKa = 6.03EE186 pKa = 4.35ARR188 pKa = 11.84MWLLKK193 pKa = 10.67NGHH196 pKa = 5.47TCDD199 pKa = 3.44NKK201 pKa = 10.17VDD203 pKa = 3.89YY204 pKa = 7.34EE205 pKa = 4.45TLTSAVTVAFMITKK219 pKa = 10.1EE220 pKa = 4.1EE221 pKa = 3.83IAFRR225 pKa = 11.84QLIKK229 pKa = 10.76NKK231 pKa = 10.34KK232 pKa = 8.36NFDD235 pKa = 3.73NMVHH239 pKa = 6.45LNKK242 pKa = 9.93TVTGNLGRR250 pKa = 11.84AAGLRR255 pKa = 11.84EE256 pKa = 4.42VKK258 pKa = 10.66SLGHH262 pKa = 5.86SFLSEE267 pKa = 3.63VHH269 pKa = 6.43LPTRR273 pKa = 11.84PIVAA277 pKa = 4.2
MM1 pKa = 7.07MLSSSVNSPVTPALSAQTPQRR22 pKa = 11.84IGGIQSKK29 pKa = 7.97RR30 pKa = 11.84QKK32 pKa = 10.4VMGYY36 pKa = 9.68LRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.06LAKK43 pKa = 9.94VPYY46 pKa = 8.86HH47 pKa = 6.36VNPALTVRR55 pKa = 11.84DD56 pKa = 3.73LHH58 pKa = 8.02VDD60 pKa = 3.49EE61 pKa = 5.54PLSKK65 pKa = 10.24EE66 pKa = 3.44QTKK69 pKa = 10.09RR70 pKa = 11.84VEE72 pKa = 3.91EE73 pKa = 4.47FIEE76 pKa = 4.3VVEE79 pKa = 4.85DD80 pKa = 3.96PKK82 pKa = 11.27SSQALKK88 pKa = 10.41DD89 pKa = 3.26NRR91 pKa = 11.84YY92 pKa = 10.25AILEE96 pKa = 4.38DD97 pKa = 4.09EE98 pKa = 4.72VKK100 pKa = 9.33HH101 pKa = 6.18TIIQMPSPTGNTYY114 pKa = 9.45QVIRR118 pKa = 11.84NKK120 pKa = 8.82TPKK123 pKa = 9.27VKK125 pKa = 10.08KK126 pKa = 10.48VKK128 pKa = 9.7IDD130 pKa = 3.6SPVMPEE136 pKa = 3.65EE137 pKa = 4.18MFPDD141 pKa = 3.12RR142 pKa = 11.84QVINRR147 pKa = 11.84QVKK150 pKa = 10.2KK151 pKa = 10.75DD152 pKa = 3.68GILAKK157 pKa = 9.74TNPSLVYY164 pKa = 9.37YY165 pKa = 10.68LRR167 pKa = 11.84TKK169 pKa = 10.68FFLKK173 pKa = 10.56FRR175 pKa = 11.84DD176 pKa = 3.6HH177 pKa = 7.85ALMQTMVHH185 pKa = 6.03EE186 pKa = 4.35ARR188 pKa = 11.84MWLLKK193 pKa = 10.67NGHH196 pKa = 5.47TCDD199 pKa = 3.44NKK201 pKa = 10.17VDD203 pKa = 3.89YY204 pKa = 7.34EE205 pKa = 4.45TLTSAVTVAFMITKK219 pKa = 10.1EE220 pKa = 4.1EE221 pKa = 3.83IAFRR225 pKa = 11.84QLIKK229 pKa = 10.76NKK231 pKa = 10.34KK232 pKa = 8.36NFDD235 pKa = 3.73NMVHH239 pKa = 6.45LNKK242 pKa = 9.93TVTGNLGRR250 pKa = 11.84AAGLRR255 pKa = 11.84EE256 pKa = 4.42VKK258 pKa = 10.66SLGHH262 pKa = 5.86SFLSEE267 pKa = 3.63VHH269 pKa = 6.43LPTRR273 pKa = 11.84PIVAA277 pKa = 4.2
Molecular weight: 31.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1113 |
277 |
425 |
371.0 |
41.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.469 ± 1.065 | 1.078 ± 0.378 |
4.762 ± 0.199 | 4.762 ± 0.625 |
3.953 ± 1.054 | 5.391 ± 0.899 |
2.156 ± 0.36 | 5.93 ± 0.751 |
7.008 ± 0.97 | 8.715 ± 0.113 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.875 ± 0.356 | 5.571 ± 0.581 |
4.762 ± 0.796 | 3.774 ± 0.393 |
6.11 ± 0.891 | 7.996 ± 0.936 |
7.457 ± 1.219 | 6.739 ± 1.719 |
0.898 ± 0.247 | 3.594 ± 0.528 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
