
candidate division MSBL1 archaeon SCGC-AAA382N08
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
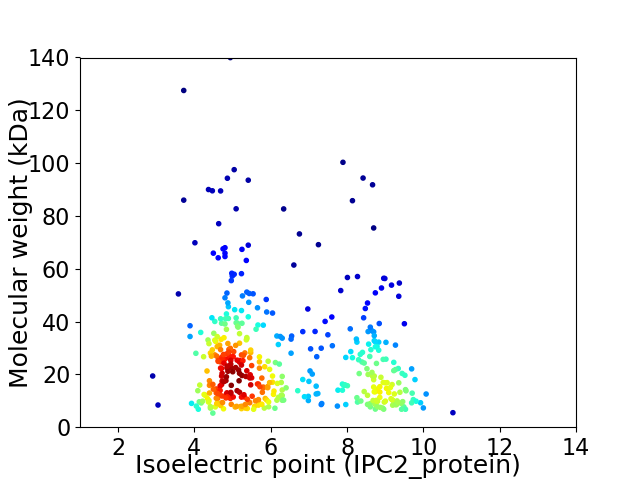
Virtual 2D-PAGE plot for 431 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
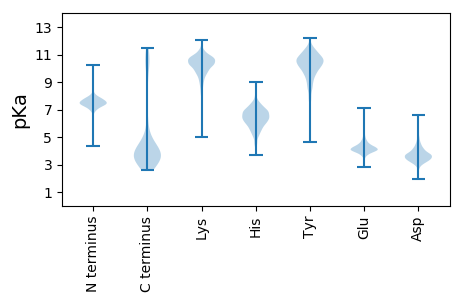
Protein with the lowest isoelectric point:
>tr|A0A133VNQ8|A0A133VNQ8_9EURY Uncharacterized protein OS=candidate division MSBL1 archaeon SCGC-AAA382N08 OX=1698285 GN=AKJ56_01915 PE=4 SV=1
MM1 pKa = 7.42NLLKK5 pKa = 10.38KK6 pKa = 10.61LSVIGLVAVFSFAMVGMALADD27 pKa = 3.67SASDD31 pKa = 3.61YY32 pKa = 10.81TSQADD37 pKa = 3.77CSDD40 pKa = 3.31AGYY43 pKa = 10.79YY44 pKa = 9.4WYY46 pKa = 9.37TGNEE50 pKa = 3.78TCYY53 pKa = 9.15EE54 pKa = 4.12TEE56 pKa = 3.93NAKK59 pKa = 10.73LSAQVSNLLEE69 pKa = 5.02RR70 pKa = 11.84IDD72 pKa = 4.51SLTATIAEE80 pKa = 4.26LTGDD84 pKa = 4.35EE85 pKa = 4.55EE86 pKa = 4.74PEE88 pKa = 4.06PATGGDD94 pKa = 3.47VPAVCEE100 pKa = 3.99GVTFDD105 pKa = 3.95RR106 pKa = 11.84ALDD109 pKa = 3.59VGMSGDD115 pKa = 5.68DD116 pKa = 3.76VLCLQTMLNQDD127 pKa = 3.81LSEE130 pKa = 5.23PIADD134 pKa = 4.48SGPGSPGNEE143 pKa = 3.22TSYY146 pKa = 10.81FGSITKK152 pKa = 10.47GGVVTFQEE160 pKa = 4.61NYY162 pKa = 10.32SEE164 pKa = 5.44DD165 pKa = 3.95VLAPWDD171 pKa = 3.78LTQGTGYY178 pKa = 10.34VGKK181 pKa = 7.18TTRR184 pKa = 11.84EE185 pKa = 4.05KK186 pKa = 11.16LNSLLDD192 pKa = 3.22QWAAEE197 pKa = 4.19EE198 pKa = 4.53KK199 pKa = 11.03APGDD203 pKa = 3.61YY204 pKa = 10.34TEE206 pKa = 5.27QSACEE211 pKa = 3.64DD212 pKa = 3.33AGYY215 pKa = 10.62YY216 pKa = 9.36WYY218 pKa = 10.17DD219 pKa = 3.61EE220 pKa = 4.38ACHH223 pKa = 5.99EE224 pKa = 4.25EE225 pKa = 4.34EE226 pKa = 4.56EE227 pKa = 4.88GEE229 pKa = 4.32EE230 pKa = 4.15VTGEE234 pKa = 4.08GLQVALADD242 pKa = 4.86DD243 pKa = 4.32NPAAATIISGTDD255 pKa = 2.95GGNAQSLVPFLKK267 pKa = 10.81VKK269 pKa = 10.83LEE271 pKa = 4.29NGDD274 pKa = 3.52NSAVDD279 pKa = 3.63VTQLDD284 pKa = 4.65FSRR287 pKa = 11.84SGISSDD293 pKa = 3.25SDD295 pKa = 2.98ISQGYY300 pKa = 8.85LFEE303 pKa = 6.71GDD305 pKa = 3.74EE306 pKa = 4.15QVAEE310 pKa = 4.02YY311 pKa = 11.03SSFNSGVMTFNDD323 pKa = 3.47SSGLFSVSANGSKK336 pKa = 10.46VVTLKK341 pKa = 10.99NDD343 pKa = 3.3IASGTASGKK352 pKa = 7.07TFKK355 pKa = 10.78FSLGEE360 pKa = 4.07ASDD363 pKa = 4.01VSSDD367 pKa = 3.23ASEE370 pKa = 4.25VSGDD374 pKa = 3.4FALNGNTMTTANVDD388 pKa = 3.73DD389 pKa = 5.15LGQITVAGTSSSPTGTVDD407 pKa = 3.22PQDD410 pKa = 3.96DD411 pKa = 4.1YY412 pKa = 12.14EE413 pKa = 4.46VFNFNLQAGQQDD425 pKa = 3.88MEE427 pKa = 4.38VQKK430 pKa = 11.24LGFTNVGSTDD440 pKa = 3.4HH441 pKa = 7.28EE442 pKa = 4.95DD443 pKa = 3.28IQDD446 pKa = 3.38FEE448 pKa = 4.97LYY450 pKa = 10.85YY451 pKa = 11.08GGTKK455 pKa = 10.26LAGPMQMASDD465 pKa = 4.02DD466 pKa = 4.08TVSFDD471 pKa = 5.62LSDD474 pKa = 3.75SPII477 pKa = 4.04
MM1 pKa = 7.42NLLKK5 pKa = 10.38KK6 pKa = 10.61LSVIGLVAVFSFAMVGMALADD27 pKa = 3.67SASDD31 pKa = 3.61YY32 pKa = 10.81TSQADD37 pKa = 3.77CSDD40 pKa = 3.31AGYY43 pKa = 10.79YY44 pKa = 9.4WYY46 pKa = 9.37TGNEE50 pKa = 3.78TCYY53 pKa = 9.15EE54 pKa = 4.12TEE56 pKa = 3.93NAKK59 pKa = 10.73LSAQVSNLLEE69 pKa = 5.02RR70 pKa = 11.84IDD72 pKa = 4.51SLTATIAEE80 pKa = 4.26LTGDD84 pKa = 4.35EE85 pKa = 4.55EE86 pKa = 4.74PEE88 pKa = 4.06PATGGDD94 pKa = 3.47VPAVCEE100 pKa = 3.99GVTFDD105 pKa = 3.95RR106 pKa = 11.84ALDD109 pKa = 3.59VGMSGDD115 pKa = 5.68DD116 pKa = 3.76VLCLQTMLNQDD127 pKa = 3.81LSEE130 pKa = 5.23PIADD134 pKa = 4.48SGPGSPGNEE143 pKa = 3.22TSYY146 pKa = 10.81FGSITKK152 pKa = 10.47GGVVTFQEE160 pKa = 4.61NYY162 pKa = 10.32SEE164 pKa = 5.44DD165 pKa = 3.95VLAPWDD171 pKa = 3.78LTQGTGYY178 pKa = 10.34VGKK181 pKa = 7.18TTRR184 pKa = 11.84EE185 pKa = 4.05KK186 pKa = 11.16LNSLLDD192 pKa = 3.22QWAAEE197 pKa = 4.19EE198 pKa = 4.53KK199 pKa = 11.03APGDD203 pKa = 3.61YY204 pKa = 10.34TEE206 pKa = 5.27QSACEE211 pKa = 3.64DD212 pKa = 3.33AGYY215 pKa = 10.62YY216 pKa = 9.36WYY218 pKa = 10.17DD219 pKa = 3.61EE220 pKa = 4.38ACHH223 pKa = 5.99EE224 pKa = 4.25EE225 pKa = 4.34EE226 pKa = 4.56EE227 pKa = 4.88GEE229 pKa = 4.32EE230 pKa = 4.15VTGEE234 pKa = 4.08GLQVALADD242 pKa = 4.86DD243 pKa = 4.32NPAAATIISGTDD255 pKa = 2.95GGNAQSLVPFLKK267 pKa = 10.81VKK269 pKa = 10.83LEE271 pKa = 4.29NGDD274 pKa = 3.52NSAVDD279 pKa = 3.63VTQLDD284 pKa = 4.65FSRR287 pKa = 11.84SGISSDD293 pKa = 3.25SDD295 pKa = 2.98ISQGYY300 pKa = 8.85LFEE303 pKa = 6.71GDD305 pKa = 3.74EE306 pKa = 4.15QVAEE310 pKa = 4.02YY311 pKa = 11.03SSFNSGVMTFNDD323 pKa = 3.47SSGLFSVSANGSKK336 pKa = 10.46VVTLKK341 pKa = 10.99NDD343 pKa = 3.3IASGTASGKK352 pKa = 7.07TFKK355 pKa = 10.78FSLGEE360 pKa = 4.07ASDD363 pKa = 4.01VSSDD367 pKa = 3.23ASEE370 pKa = 4.25VSGDD374 pKa = 3.4FALNGNTMTTANVDD388 pKa = 3.73DD389 pKa = 5.15LGQITVAGTSSSPTGTVDD407 pKa = 3.22PQDD410 pKa = 3.96DD411 pKa = 4.1YY412 pKa = 12.14EE413 pKa = 4.46VFNFNLQAGQQDD425 pKa = 3.88MEE427 pKa = 4.38VQKK430 pKa = 11.24LGFTNVGSTDD440 pKa = 3.4HH441 pKa = 7.28EE442 pKa = 4.95DD443 pKa = 3.28IQDD446 pKa = 3.38FEE448 pKa = 4.97LYY450 pKa = 10.85YY451 pKa = 11.08GGTKK455 pKa = 10.26LAGPMQMASDD465 pKa = 4.02DD466 pKa = 4.08TVSFDD471 pKa = 5.62LSDD474 pKa = 3.75SPII477 pKa = 4.04
Molecular weight: 50.45 kDa
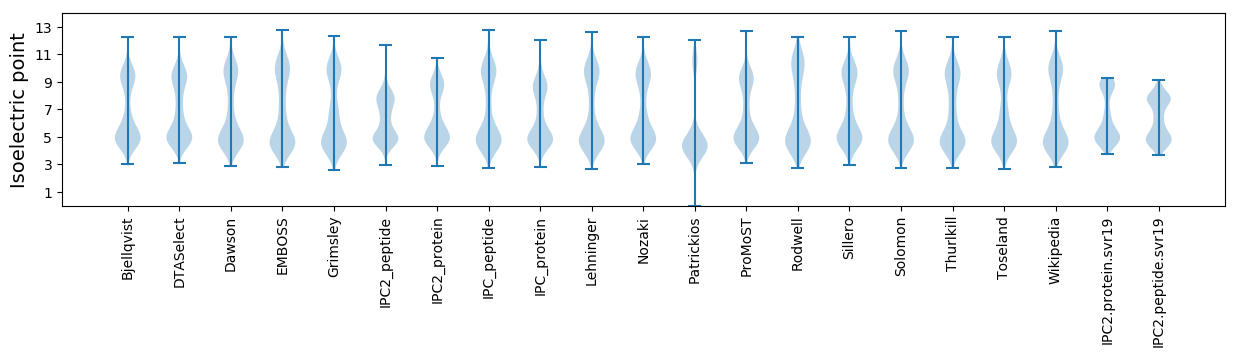
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133VPE1|A0A133VPE1_9EURY Uncharacterized protein OS=candidate division MSBL1 archaeon SCGC-AAA382N08 OX=1698285 GN=AKJ56_01585 PE=4 SV=1
MM1 pKa = 7.32YY2 pKa = 8.87VTQTNFSARR11 pKa = 11.84EE12 pKa = 3.77LLKK15 pKa = 10.89VLFRR19 pKa = 11.84NRR21 pKa = 11.84FRR23 pKa = 11.84IVNRR27 pKa = 11.84TGSNVKK33 pKa = 9.87LRR35 pKa = 11.84YY36 pKa = 8.11EE37 pKa = 4.56HH38 pKa = 6.9PTNDD42 pKa = 3.34DD43 pKa = 3.18NNRR46 pKa = 11.84IVIVPMHH53 pKa = 6.98DD54 pKa = 3.78SIKK57 pKa = 9.91TGTLRR62 pKa = 11.84SIANQAGAKK71 pKa = 9.49NFQKK75 pKa = 10.56IKK77 pKa = 10.86DD78 pKa = 3.96WINQNII84 pKa = 3.45
MM1 pKa = 7.32YY2 pKa = 8.87VTQTNFSARR11 pKa = 11.84EE12 pKa = 3.77LLKK15 pKa = 10.89VLFRR19 pKa = 11.84NRR21 pKa = 11.84FRR23 pKa = 11.84IVNRR27 pKa = 11.84TGSNVKK33 pKa = 9.87LRR35 pKa = 11.84YY36 pKa = 8.11EE37 pKa = 4.56HH38 pKa = 6.9PTNDD42 pKa = 3.34DD43 pKa = 3.18NNRR46 pKa = 11.84IVIVPMHH53 pKa = 6.98DD54 pKa = 3.78SIKK57 pKa = 9.91TGTLRR62 pKa = 11.84SIANQAGAKK71 pKa = 9.49NFQKK75 pKa = 10.56IKK77 pKa = 10.86DD78 pKa = 3.96WINQNII84 pKa = 3.45
Molecular weight: 9.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
101451 |
45 |
1209 |
235.4 |
26.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.618 ± 0.119 | 0.914 ± 0.052 |
5.817 ± 0.11 | 9.934 ± 0.203 |
4.047 ± 0.097 | 7.001 ± 0.134 |
1.461 ± 0.051 | 6.959 ± 0.112 |
8.603 ± 0.197 | 8.899 ± 0.153 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.06 | 4.589 ± 0.115 |
3.656 ± 0.062 | 2.965 ± 0.106 |
5.009 ± 0.117 | 6.528 ± 0.14 |
4.928 ± 0.124 | 6.508 ± 0.107 |
1.159 ± 0.044 | 3.27 ± 0.084 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
