
Blastococcus sp. TF02A-30
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
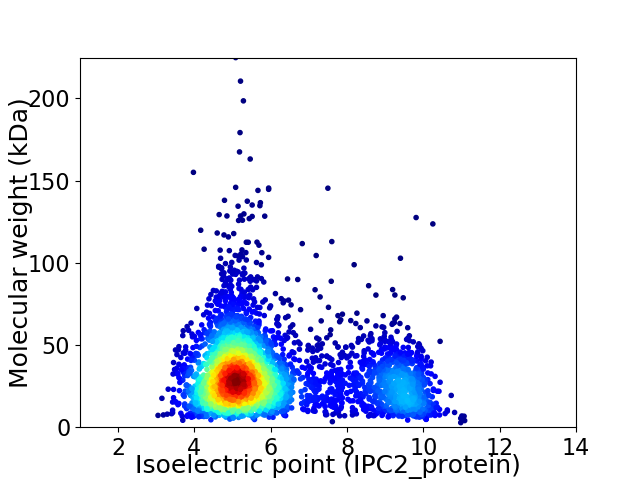
Virtual 2D-PAGE plot for 3930 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A367ALH2|A0A367ALH2_9ACTN NADP-dependent oxidoreductase OS=Blastococcus sp. TF02A-30 OX=2250580 GN=DQ241_08025 PE=4 SV=1
MM1 pKa = 7.72KK2 pKa = 9.25LTKK5 pKa = 10.11RR6 pKa = 11.84RR7 pKa = 11.84GAFVATALSAALVLSACGGGDD28 pKa = 4.33DD29 pKa = 4.44EE30 pKa = 6.67GGDD33 pKa = 3.79GGSASGGGEE42 pKa = 3.33FSIYY46 pKa = 10.23IGDD49 pKa = 4.11PEE51 pKa = 4.44NPLIPGNTTEE61 pKa = 4.32TEE63 pKa = 4.04GGQVVDD69 pKa = 4.19TLWTGLVTYY78 pKa = 10.28DD79 pKa = 4.41HH80 pKa = 6.69EE81 pKa = 4.63TSEE84 pKa = 4.21ATFDD88 pKa = 3.76GVAEE92 pKa = 4.58SIEE95 pKa = 4.78SDD97 pKa = 4.19DD98 pKa = 3.55QTTWTVTLKK107 pKa = 10.95DD108 pKa = 2.87GWTFHH113 pKa = 7.74DD114 pKa = 4.11GTPVNAEE121 pKa = 3.95SFVNAWNWNAYY132 pKa = 9.45SPNAAGNSYY141 pKa = 10.31FFANIEE147 pKa = 4.49GYY149 pKa = 10.57DD150 pKa = 3.62EE151 pKa = 4.4LQAEE155 pKa = 4.6EE156 pKa = 5.11GGTPAADD163 pKa = 3.22AMSGLTVVDD172 pKa = 4.43DD173 pKa = 3.81LTFEE177 pKa = 4.58VKK179 pKa = 10.67LSAPYY184 pKa = 9.87AQWPTTVGYY193 pKa = 7.89TAFFPLPEE201 pKa = 4.67AFFDD205 pKa = 4.02DD206 pKa = 4.57PEE208 pKa = 5.77AFGSQPIGNGPFQAEE223 pKa = 4.13EE224 pKa = 4.18PFIEE228 pKa = 4.74GQGITLTKK236 pKa = 10.34FEE238 pKa = 5.52DD239 pKa = 4.07YY240 pKa = 10.93AGEE243 pKa = 4.67DD244 pKa = 3.6EE245 pKa = 4.6AQADD249 pKa = 3.67AVRR252 pKa = 11.84YY253 pKa = 9.09VVYY256 pKa = 10.47TEE258 pKa = 5.6LNTAYY263 pKa = 9.57TDD265 pKa = 3.93LQGGTLDD272 pKa = 4.89IVDD275 pKa = 4.86TLPPDD280 pKa = 5.12AIASAEE286 pKa = 4.15GEE288 pKa = 4.0FGDD291 pKa = 5.31RR292 pKa = 11.84YY293 pKa = 10.52IEE295 pKa = 4.07TAQGDD300 pKa = 3.84ITSLGFPTYY309 pKa = 10.13DD310 pKa = 4.1EE311 pKa = 5.18RR312 pKa = 11.84FADD315 pKa = 3.77PNVRR319 pKa = 11.84KK320 pKa = 9.59AFSMAIDD327 pKa = 3.64RR328 pKa = 11.84QSISDD333 pKa = 4.15AIFNGTRR340 pKa = 11.84TPATSYY346 pKa = 10.67VSPVVDD352 pKa = 4.93GYY354 pKa = 11.58RR355 pKa = 11.84DD356 pKa = 3.78DD357 pKa = 4.76ACEE360 pKa = 3.97ACEE363 pKa = 5.31LDD365 pKa = 3.31VDD367 pKa = 4.07EE368 pKa = 5.97ANRR371 pKa = 11.84LLDD374 pKa = 3.33EE375 pKa = 5.15AGFDD379 pKa = 3.36RR380 pKa = 11.84TQPVDD385 pKa = 2.88LWFNAGGGHH394 pKa = 5.67EE395 pKa = 4.32EE396 pKa = 3.85WMQAVGNQIRR406 pKa = 11.84TNLGVEE412 pKa = 3.95YY413 pKa = 10.09QLRR416 pKa = 11.84GDD418 pKa = 3.92LQFAEE423 pKa = 4.45YY424 pKa = 10.73LPLQDD429 pKa = 3.88AQGMTGPFRR438 pKa = 11.84SGWIMDD444 pKa = 3.89YY445 pKa = 11.05PVMEE449 pKa = 4.64NFLGPLYY456 pKa = 9.91STAALPPAGSNVTFYY471 pKa = 11.55SNPEE475 pKa = 3.17FDD477 pKa = 4.29AALEE481 pKa = 4.48AGNSAASIDD490 pKa = 4.02EE491 pKa = 5.35AITAYY496 pKa = 10.51QEE498 pKa = 4.42AEE500 pKa = 4.59DD501 pKa = 4.51ILLEE505 pKa = 4.54DD506 pKa = 5.34FPAAPLFYY514 pKa = 10.52RR515 pKa = 11.84LNQGAHH521 pKa = 5.34SEE523 pKa = 4.19RR524 pKa = 11.84VSNVIINAFGRR535 pKa = 11.84IDD537 pKa = 3.43AAAVTVNDD545 pKa = 3.78
MM1 pKa = 7.72KK2 pKa = 9.25LTKK5 pKa = 10.11RR6 pKa = 11.84RR7 pKa = 11.84GAFVATALSAALVLSACGGGDD28 pKa = 4.33DD29 pKa = 4.44EE30 pKa = 6.67GGDD33 pKa = 3.79GGSASGGGEE42 pKa = 3.33FSIYY46 pKa = 10.23IGDD49 pKa = 4.11PEE51 pKa = 4.44NPLIPGNTTEE61 pKa = 4.32TEE63 pKa = 4.04GGQVVDD69 pKa = 4.19TLWTGLVTYY78 pKa = 10.28DD79 pKa = 4.41HH80 pKa = 6.69EE81 pKa = 4.63TSEE84 pKa = 4.21ATFDD88 pKa = 3.76GVAEE92 pKa = 4.58SIEE95 pKa = 4.78SDD97 pKa = 4.19DD98 pKa = 3.55QTTWTVTLKK107 pKa = 10.95DD108 pKa = 2.87GWTFHH113 pKa = 7.74DD114 pKa = 4.11GTPVNAEE121 pKa = 3.95SFVNAWNWNAYY132 pKa = 9.45SPNAAGNSYY141 pKa = 10.31FFANIEE147 pKa = 4.49GYY149 pKa = 10.57DD150 pKa = 3.62EE151 pKa = 4.4LQAEE155 pKa = 4.6EE156 pKa = 5.11GGTPAADD163 pKa = 3.22AMSGLTVVDD172 pKa = 4.43DD173 pKa = 3.81LTFEE177 pKa = 4.58VKK179 pKa = 10.67LSAPYY184 pKa = 9.87AQWPTTVGYY193 pKa = 7.89TAFFPLPEE201 pKa = 4.67AFFDD205 pKa = 4.02DD206 pKa = 4.57PEE208 pKa = 5.77AFGSQPIGNGPFQAEE223 pKa = 4.13EE224 pKa = 4.18PFIEE228 pKa = 4.74GQGITLTKK236 pKa = 10.34FEE238 pKa = 5.52DD239 pKa = 4.07YY240 pKa = 10.93AGEE243 pKa = 4.67DD244 pKa = 3.6EE245 pKa = 4.6AQADD249 pKa = 3.67AVRR252 pKa = 11.84YY253 pKa = 9.09VVYY256 pKa = 10.47TEE258 pKa = 5.6LNTAYY263 pKa = 9.57TDD265 pKa = 3.93LQGGTLDD272 pKa = 4.89IVDD275 pKa = 4.86TLPPDD280 pKa = 5.12AIASAEE286 pKa = 4.15GEE288 pKa = 4.0FGDD291 pKa = 5.31RR292 pKa = 11.84YY293 pKa = 10.52IEE295 pKa = 4.07TAQGDD300 pKa = 3.84ITSLGFPTYY309 pKa = 10.13DD310 pKa = 4.1EE311 pKa = 5.18RR312 pKa = 11.84FADD315 pKa = 3.77PNVRR319 pKa = 11.84KK320 pKa = 9.59AFSMAIDD327 pKa = 3.64RR328 pKa = 11.84QSISDD333 pKa = 4.15AIFNGTRR340 pKa = 11.84TPATSYY346 pKa = 10.67VSPVVDD352 pKa = 4.93GYY354 pKa = 11.58RR355 pKa = 11.84DD356 pKa = 3.78DD357 pKa = 4.76ACEE360 pKa = 3.97ACEE363 pKa = 5.31LDD365 pKa = 3.31VDD367 pKa = 4.07EE368 pKa = 5.97ANRR371 pKa = 11.84LLDD374 pKa = 3.33EE375 pKa = 5.15AGFDD379 pKa = 3.36RR380 pKa = 11.84TQPVDD385 pKa = 2.88LWFNAGGGHH394 pKa = 5.67EE395 pKa = 4.32EE396 pKa = 3.85WMQAVGNQIRR406 pKa = 11.84TNLGVEE412 pKa = 3.95YY413 pKa = 10.09QLRR416 pKa = 11.84GDD418 pKa = 3.92LQFAEE423 pKa = 4.45YY424 pKa = 10.73LPLQDD429 pKa = 3.88AQGMTGPFRR438 pKa = 11.84SGWIMDD444 pKa = 3.89YY445 pKa = 11.05PVMEE449 pKa = 4.64NFLGPLYY456 pKa = 9.91STAALPPAGSNVTFYY471 pKa = 11.55SNPEE475 pKa = 3.17FDD477 pKa = 4.29AALEE481 pKa = 4.48AGNSAASIDD490 pKa = 4.02EE491 pKa = 5.35AITAYY496 pKa = 10.51QEE498 pKa = 4.42AEE500 pKa = 4.59DD501 pKa = 4.51ILLEE505 pKa = 4.54DD506 pKa = 5.34FPAAPLFYY514 pKa = 10.52RR515 pKa = 11.84LNQGAHH521 pKa = 5.34SEE523 pKa = 4.19RR524 pKa = 11.84VSNVIINAFGRR535 pKa = 11.84IDD537 pKa = 3.43AAAVTVNDD545 pKa = 3.78
Molecular weight: 58.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A367AV44|A0A367AV44_9ACTN Uncharacterized protein OS=Blastococcus sp. TF02A-30 OX=2250580 GN=DQ241_06275 PE=4 SV=1
MM1 pKa = 6.81PTRR4 pKa = 11.84WGRR7 pKa = 11.84GRR9 pKa = 11.84GGPPGRR15 pKa = 11.84AAPSGSAPGGPCGAPRR31 pKa = 11.84RR32 pKa = 11.84AAAAGARR39 pKa = 11.84PGRR42 pKa = 11.84RR43 pKa = 11.84GPGRR47 pKa = 11.84RR48 pKa = 11.84SGPPASRR55 pKa = 11.84GPARR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84GRR63 pKa = 11.84PGALPP68 pKa = 3.66
MM1 pKa = 6.81PTRR4 pKa = 11.84WGRR7 pKa = 11.84GRR9 pKa = 11.84GGPPGRR15 pKa = 11.84AAPSGSAPGGPCGAPRR31 pKa = 11.84RR32 pKa = 11.84AAAAGARR39 pKa = 11.84PGRR42 pKa = 11.84RR43 pKa = 11.84GPGRR47 pKa = 11.84RR48 pKa = 11.84SGPPASRR55 pKa = 11.84GPARR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84GRR63 pKa = 11.84PGALPP68 pKa = 3.66
Molecular weight: 6.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1241105 |
24 |
2049 |
315.8 |
33.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.43 ± 0.061 | 0.66 ± 0.011 |
6.228 ± 0.033 | 5.759 ± 0.041 |
2.574 ± 0.022 | 9.67 ± 0.042 |
1.975 ± 0.018 | 2.817 ± 0.029 |
1.435 ± 0.023 | 10.799 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.591 ± 0.013 | 1.424 ± 0.021 |
6.182 ± 0.036 | 2.726 ± 0.021 |
8.22 ± 0.049 | 4.698 ± 0.026 |
5.81 ± 0.029 | 9.817 ± 0.04 |
1.477 ± 0.017 | 1.709 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
