
Eubacterium sp. 14-2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; unclassified Eubacterium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
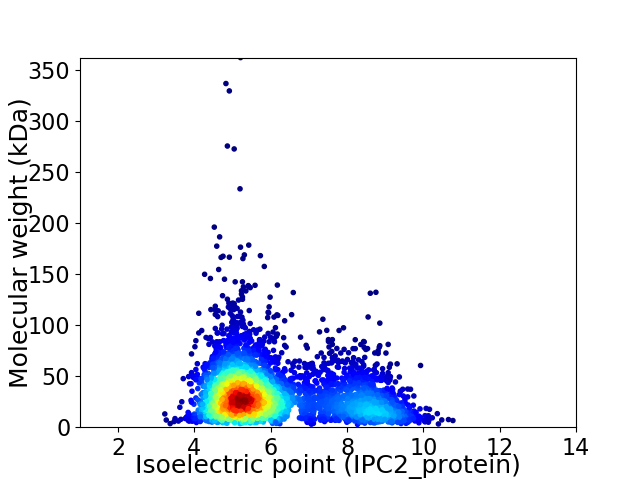
Virtual 2D-PAGE plot for 3773 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
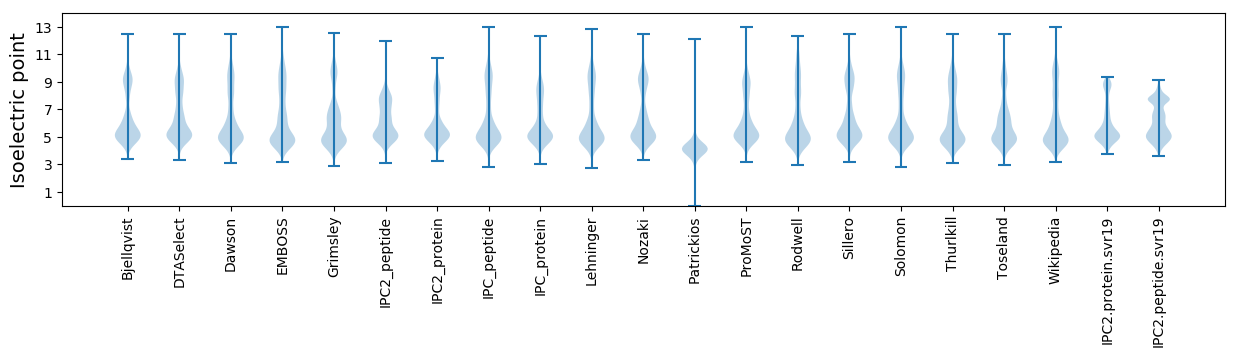
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S0J1P8|S0J1P8_9FIRM Glyco_trans_2-like domain-containing protein OS=Eubacterium sp. 14-2 OX=1235790 GN=C805_03404 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.37KK3 pKa = 10.2KK4 pKa = 10.48LLAALLCVAMSASMLAGCGNNSGGNNSANAGDD36 pKa = 4.64DD37 pKa = 3.7KK38 pKa = 11.75QEE40 pKa = 4.31TGDD43 pKa = 4.46DD44 pKa = 3.72AAGDD48 pKa = 3.89DD49 pKa = 5.15AEE51 pKa = 6.14AGDD54 pKa = 4.44DD55 pKa = 4.87AEE57 pKa = 6.21AGDD60 pKa = 4.43DD61 pKa = 3.94AAAGDD66 pKa = 4.57EE67 pKa = 4.14NTLTVWCWDD76 pKa = 3.48PKK78 pKa = 9.91FNIYY82 pKa = 11.02AMEE85 pKa = 4.21EE86 pKa = 3.89AGKK89 pKa = 10.27LYY91 pKa = 10.58QADD94 pKa = 3.47HH95 pKa = 7.26PEE97 pKa = 3.88FNINVVEE104 pKa = 4.43TPWDD108 pKa = 4.73DD109 pKa = 3.35IQKK112 pKa = 10.79KK113 pKa = 8.07LTTAATGGALDD124 pKa = 4.0TLPDD128 pKa = 3.33IMLVQDD134 pKa = 3.7NAFQKK139 pKa = 10.82NVISFPEE146 pKa = 4.33AFTDD150 pKa = 3.9LTDD153 pKa = 3.59SGVDD157 pKa = 3.74FSQFGEE163 pKa = 4.06AKK165 pKa = 8.02TAYY168 pKa = 10.1SVVEE172 pKa = 4.01GKK174 pKa = 10.46NYY176 pKa = 10.08GVPFDD181 pKa = 4.06NGTVVACYY189 pKa = 8.94RR190 pKa = 11.84TDD192 pKa = 3.32VLEE195 pKa = 4.31EE196 pKa = 4.04AGFTVDD202 pKa = 5.89DD203 pKa = 4.27FTDD206 pKa = 3.75ITWDD210 pKa = 3.34EE211 pKa = 4.09YY212 pKa = 11.31LEE214 pKa = 4.3KK215 pKa = 10.96GKK217 pKa = 10.78VVLEE221 pKa = 4.15KK222 pKa = 10.13TGQPLLSCQSTEE234 pKa = 3.74SDD236 pKa = 4.16VIMMMLQSCGASLFDD251 pKa = 6.17DD252 pKa = 4.01EE253 pKa = 5.94GNPNMVGNDD262 pKa = 3.35SLKK265 pKa = 10.83KK266 pKa = 10.77VMEE269 pKa = 4.7TYY271 pKa = 9.98QQLVEE276 pKa = 4.19TGVCKK281 pKa = 10.46LVNSWDD287 pKa = 4.1EE288 pKa = 4.08YY289 pKa = 10.34VQTLTTGTVAGTINGCWIMASIQSAEE315 pKa = 4.11DD316 pKa = 3.54QSGKK320 pKa = 9.23WALTNMPSLVGVEE333 pKa = 4.18GATNYY338 pKa = 10.69SNNGGSSWAITSNCKK353 pKa = 10.19NMEE356 pKa = 4.14LAKK359 pKa = 10.78DD360 pKa = 3.76FLKK363 pKa = 10.13STFAGSVEE371 pKa = 4.42LYY373 pKa = 10.98EE374 pKa = 5.11NILPSSGALATYY386 pKa = 10.28LPAGDD391 pKa = 3.8SDD393 pKa = 4.89VYY395 pKa = 11.35AEE397 pKa = 4.09PSEE400 pKa = 4.89FYY402 pKa = 11.11GGDD405 pKa = 3.22AVFKK409 pKa = 10.69KK410 pKa = 8.7ITEE413 pKa = 4.27YY414 pKa = 11.16ASKK417 pKa = 10.7CPSNNTGVYY426 pKa = 9.86YY427 pKa = 10.68YY428 pKa = 9.61EE429 pKa = 4.5ARR431 pKa = 11.84DD432 pKa = 4.04AIATAIQNVVGGSDD446 pKa = 3.1IDD448 pKa = 4.05SEE450 pKa = 4.98LEE452 pKa = 3.78IAQGTVEE459 pKa = 5.08GLMEE463 pKa = 3.89QQ464 pKa = 3.7
MM1 pKa = 7.48KK2 pKa = 10.37KK3 pKa = 10.2KK4 pKa = 10.48LLAALLCVAMSASMLAGCGNNSGGNNSANAGDD36 pKa = 4.64DD37 pKa = 3.7KK38 pKa = 11.75QEE40 pKa = 4.31TGDD43 pKa = 4.46DD44 pKa = 3.72AAGDD48 pKa = 3.89DD49 pKa = 5.15AEE51 pKa = 6.14AGDD54 pKa = 4.44DD55 pKa = 4.87AEE57 pKa = 6.21AGDD60 pKa = 4.43DD61 pKa = 3.94AAAGDD66 pKa = 4.57EE67 pKa = 4.14NTLTVWCWDD76 pKa = 3.48PKK78 pKa = 9.91FNIYY82 pKa = 11.02AMEE85 pKa = 4.21EE86 pKa = 3.89AGKK89 pKa = 10.27LYY91 pKa = 10.58QADD94 pKa = 3.47HH95 pKa = 7.26PEE97 pKa = 3.88FNINVVEE104 pKa = 4.43TPWDD108 pKa = 4.73DD109 pKa = 3.35IQKK112 pKa = 10.79KK113 pKa = 8.07LTTAATGGALDD124 pKa = 4.0TLPDD128 pKa = 3.33IMLVQDD134 pKa = 3.7NAFQKK139 pKa = 10.82NVISFPEE146 pKa = 4.33AFTDD150 pKa = 3.9LTDD153 pKa = 3.59SGVDD157 pKa = 3.74FSQFGEE163 pKa = 4.06AKK165 pKa = 8.02TAYY168 pKa = 10.1SVVEE172 pKa = 4.01GKK174 pKa = 10.46NYY176 pKa = 10.08GVPFDD181 pKa = 4.06NGTVVACYY189 pKa = 8.94RR190 pKa = 11.84TDD192 pKa = 3.32VLEE195 pKa = 4.31EE196 pKa = 4.04AGFTVDD202 pKa = 5.89DD203 pKa = 4.27FTDD206 pKa = 3.75ITWDD210 pKa = 3.34EE211 pKa = 4.09YY212 pKa = 11.31LEE214 pKa = 4.3KK215 pKa = 10.96GKK217 pKa = 10.78VVLEE221 pKa = 4.15KK222 pKa = 10.13TGQPLLSCQSTEE234 pKa = 3.74SDD236 pKa = 4.16VIMMMLQSCGASLFDD251 pKa = 6.17DD252 pKa = 4.01EE253 pKa = 5.94GNPNMVGNDD262 pKa = 3.35SLKK265 pKa = 10.83KK266 pKa = 10.77VMEE269 pKa = 4.7TYY271 pKa = 9.98QQLVEE276 pKa = 4.19TGVCKK281 pKa = 10.46LVNSWDD287 pKa = 4.1EE288 pKa = 4.08YY289 pKa = 10.34VQTLTTGTVAGTINGCWIMASIQSAEE315 pKa = 4.11DD316 pKa = 3.54QSGKK320 pKa = 9.23WALTNMPSLVGVEE333 pKa = 4.18GATNYY338 pKa = 10.69SNNGGSSWAITSNCKK353 pKa = 10.19NMEE356 pKa = 4.14LAKK359 pKa = 10.78DD360 pKa = 3.76FLKK363 pKa = 10.13STFAGSVEE371 pKa = 4.42LYY373 pKa = 10.98EE374 pKa = 5.11NILPSSGALATYY386 pKa = 10.28LPAGDD391 pKa = 3.8SDD393 pKa = 4.89VYY395 pKa = 11.35AEE397 pKa = 4.09PSEE400 pKa = 4.89FYY402 pKa = 11.11GGDD405 pKa = 3.22AVFKK409 pKa = 10.69KK410 pKa = 8.7ITEE413 pKa = 4.27YY414 pKa = 11.16ASKK417 pKa = 10.7CPSNNTGVYY426 pKa = 9.86YY427 pKa = 10.68YY428 pKa = 9.61EE429 pKa = 4.5ARR431 pKa = 11.84DD432 pKa = 4.04AIATAIQNVVGGSDD446 pKa = 3.1IDD448 pKa = 4.05SEE450 pKa = 4.98LEE452 pKa = 3.78IAQGTVEE459 pKa = 5.08GLMEE463 pKa = 3.89QQ464 pKa = 3.7
Molecular weight: 49.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S0JJ38|S0JJ38_9FIRM HTH cro/C1-type domain-containing protein OS=Eubacterium sp. 14-2 OX=1235790 GN=C805_01104 PE=4 SV=1
MM1 pKa = 7.84AGMVLRR7 pKa = 11.84KK8 pKa = 8.44TGVVLLTILKK18 pKa = 9.45WILQAALVALKK29 pKa = 10.49LVLGIAKK36 pKa = 10.15LFLLLLSLVMRR47 pKa = 11.84IVLTMIGVSVGRR59 pKa = 11.84RR60 pKa = 3.23
MM1 pKa = 7.84AGMVLRR7 pKa = 11.84KK8 pKa = 8.44TGVVLLTILKK18 pKa = 9.45WILQAALVALKK29 pKa = 10.49LVLGIAKK36 pKa = 10.15LFLLLLSLVMRR47 pKa = 11.84IVLTMIGVSVGRR59 pKa = 11.84RR60 pKa = 3.23
Molecular weight: 6.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1163219 |
26 |
3240 |
308.3 |
34.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.176 ± 0.045 | 1.515 ± 0.019 |
5.234 ± 0.026 | 8.517 ± 0.047 |
4.072 ± 0.025 | 7.143 ± 0.039 |
1.729 ± 0.019 | 6.969 ± 0.035 |
6.621 ± 0.039 | 8.965 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.146 ± 0.022 | 4.273 ± 0.029 |
3.25 ± 0.026 | 3.668 ± 0.029 |
4.962 ± 0.039 | 5.874 ± 0.037 |
5.21 ± 0.038 | 6.673 ± 0.032 |
0.93 ± 0.014 | 4.073 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
