
Alphacoronavirus BtMs-AlphaCoV/GS2013
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Decacovirus; Rhinolophus ferrumequinum alphacoronavirus HuB-2013
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
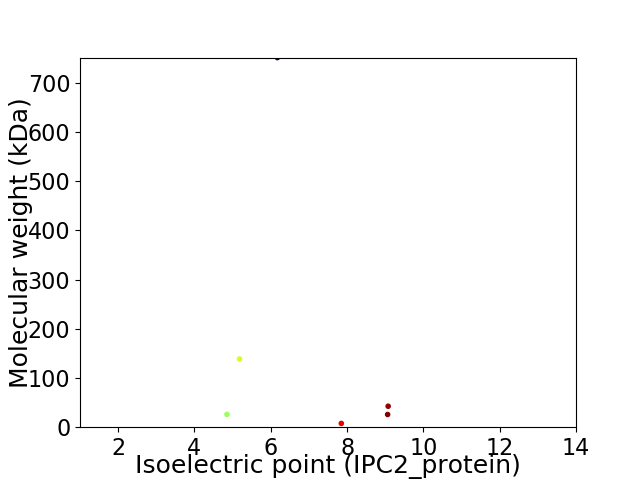
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U1WJX0|A0A0U1WJX0_9ALPC Small envelope protein OS=Alphacoronavirus BtMs-AlphaCoV/GS2013 OX=1503290 PE=4 SV=1
MM1 pKa = 7.43FLGLFEE7 pKa = 5.52SSMEE11 pKa = 3.81AAIANGFNIPPNEE24 pKa = 3.98VEE26 pKa = 4.91EE27 pKa = 4.22ILDD30 pKa = 3.8VIKK33 pKa = 10.07PARR36 pKa = 11.84QTISIASFLLSSIFVTWFALYY57 pKa = 10.07KK58 pKa = 10.48ASCFRR63 pKa = 11.84TNCALFALRR72 pKa = 11.84ICTLLLYY79 pKa = 10.09TPILVFFNSYY89 pKa = 9.21IDD91 pKa = 3.77AVVVACSLLLRR102 pKa = 11.84FAYY105 pKa = 10.25LGYY108 pKa = 10.72YY109 pKa = 8.69SYY111 pKa = 11.07KK112 pKa = 9.51YY113 pKa = 10.47KK114 pKa = 10.83SFSFLVLNTSKK125 pKa = 10.89VAFVNGKK132 pKa = 8.15FWYY135 pKa = 9.45YY136 pKa = 10.92DD137 pKa = 3.33EE138 pKa = 4.84SPYY141 pKa = 10.91VVLYY145 pKa = 11.05GGDD148 pKa = 3.1HH149 pKa = 6.05HH150 pKa = 7.05VQFGEE155 pKa = 3.98YY156 pKa = 8.55MIPFADD162 pKa = 3.86SNEE165 pKa = 4.27LYY167 pKa = 10.54VALRR171 pKa = 11.84GTTEE175 pKa = 4.3DD176 pKa = 4.5DD177 pKa = 3.63VPLSRR182 pKa = 11.84KK183 pKa = 9.67VEE185 pKa = 3.94MMNGAFIYY193 pKa = 10.22IFARR197 pKa = 11.84EE198 pKa = 4.11PCVGIVNMSFKK209 pKa = 9.62EE210 pKa = 4.29TQLDD214 pKa = 3.63EE215 pKa = 4.99DD216 pKa = 4.01LTSITLHH223 pKa = 6.13NVDD226 pKa = 3.96ISQQ229 pKa = 3.45
MM1 pKa = 7.43FLGLFEE7 pKa = 5.52SSMEE11 pKa = 3.81AAIANGFNIPPNEE24 pKa = 3.98VEE26 pKa = 4.91EE27 pKa = 4.22ILDD30 pKa = 3.8VIKK33 pKa = 10.07PARR36 pKa = 11.84QTISIASFLLSSIFVTWFALYY57 pKa = 10.07KK58 pKa = 10.48ASCFRR63 pKa = 11.84TNCALFALRR72 pKa = 11.84ICTLLLYY79 pKa = 10.09TPILVFFNSYY89 pKa = 9.21IDD91 pKa = 3.77AVVVACSLLLRR102 pKa = 11.84FAYY105 pKa = 10.25LGYY108 pKa = 10.72YY109 pKa = 8.69SYY111 pKa = 11.07KK112 pKa = 9.51YY113 pKa = 10.47KK114 pKa = 10.83SFSFLVLNTSKK125 pKa = 10.89VAFVNGKK132 pKa = 8.15FWYY135 pKa = 9.45YY136 pKa = 10.92DD137 pKa = 3.33EE138 pKa = 4.84SPYY141 pKa = 10.91VVLYY145 pKa = 11.05GGDD148 pKa = 3.1HH149 pKa = 6.05HH150 pKa = 7.05VQFGEE155 pKa = 3.98YY156 pKa = 8.55MIPFADD162 pKa = 3.86SNEE165 pKa = 4.27LYY167 pKa = 10.54VALRR171 pKa = 11.84GTTEE175 pKa = 4.3DD176 pKa = 4.5DD177 pKa = 3.63VPLSRR182 pKa = 11.84KK183 pKa = 9.67VEE185 pKa = 3.94MMNGAFIYY193 pKa = 10.22IFARR197 pKa = 11.84EE198 pKa = 4.11PCVGIVNMSFKK209 pKa = 9.62EE210 pKa = 4.29TQLDD214 pKa = 3.63EE215 pKa = 4.99DD216 pKa = 4.01LTSITLHH223 pKa = 6.13NVDD226 pKa = 3.96ISQQ229 pKa = 3.45
Molecular weight: 26.06 kDa
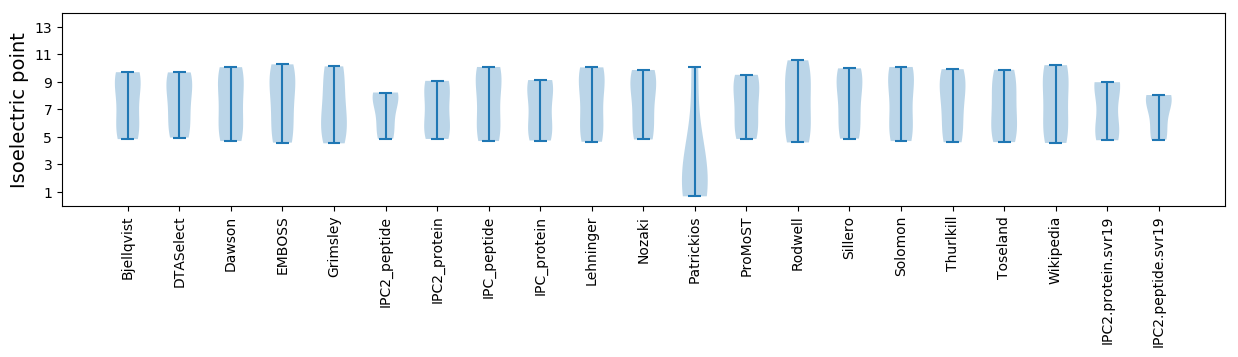
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U1WHF8|A0A0U1WHF8_9ALPC Membrane protein OS=Alphacoronavirus BtMs-AlphaCoV/GS2013 OX=1503290 GN=M PE=3 SV=1
MM1 pKa = 7.6ASVSFQEE8 pKa = 4.47EE9 pKa = 3.91KK10 pKa = 10.28RR11 pKa = 11.84GRR13 pKa = 11.84SGKK16 pKa = 10.09VPLSYY21 pKa = 10.71YY22 pKa = 10.67NPVIVSGDD30 pKa = 3.33KK31 pKa = 10.81PFWKK35 pKa = 10.34VMPNNAVPKK44 pKa = 10.6GKK46 pKa = 10.41GNKK49 pKa = 8.87DD50 pKa = 3.28QQIGYY55 pKa = 8.97WNEE58 pKa = 3.5QPRR61 pKa = 11.84YY62 pKa = 9.28RR63 pKa = 11.84MVRR66 pKa = 11.84GTRR69 pKa = 11.84KK70 pKa = 9.91DD71 pKa = 4.11LPSKK75 pKa = 8.43WHH77 pKa = 6.58FYY79 pKa = 10.71YY80 pKa = 10.91LGTGPHH86 pKa = 6.67AEE88 pKa = 3.88AKK90 pKa = 10.13FRR92 pKa = 11.84TRR94 pKa = 11.84TDD96 pKa = 2.94GVFWVAVQGSKK107 pKa = 9.39TEE109 pKa = 3.99PTGLGTRR116 pKa = 11.84KK117 pKa = 9.96RR118 pKa = 11.84NAEE121 pKa = 3.92LVNPEE126 pKa = 4.35FAIQLPAAIEE136 pKa = 4.03IQEE139 pKa = 4.22NTVSRR144 pKa = 11.84GNSRR148 pKa = 11.84NQSSNRR154 pKa = 11.84DD155 pKa = 3.21RR156 pKa = 11.84SQSGNRR162 pKa = 11.84SQNDD166 pKa = 3.31QKK168 pKa = 11.2QGNQNQKK175 pKa = 10.14SRR177 pKa = 11.84SNSQSRR183 pKa = 11.84KK184 pKa = 9.77GNQNSNDD191 pKa = 3.6SAVDD195 pKa = 2.96IVAAVKK201 pKa = 10.1QALKK205 pKa = 10.54EE206 pKa = 3.9LGVTNEE212 pKa = 4.09SNKK215 pKa = 9.98KK216 pKa = 9.45GKK218 pKa = 10.01NSGTTTPKK226 pKa = 9.52EE227 pKa = 4.05QRR229 pKa = 11.84SKK231 pKa = 11.36SPVRR235 pKa = 11.84SPTVQKK241 pKa = 10.58KK242 pKa = 7.53QLEE245 pKa = 4.23RR246 pKa = 11.84PPWKK250 pKa = 9.82RR251 pKa = 11.84VPNSTEE257 pKa = 4.38DD258 pKa = 3.29VTKK261 pKa = 10.8CFGVRR266 pKa = 11.84DD267 pKa = 3.94TYY269 pKa = 11.62RR270 pKa = 11.84NFGDD274 pKa = 3.78ADD276 pKa = 3.87LVRR279 pKa = 11.84NGIDD283 pKa = 3.11AKK285 pKa = 10.61HH286 pKa = 5.49YY287 pKa = 7.59PQLAEE292 pKa = 4.62LVPTPAAILFGGEE305 pKa = 4.17VVTKK309 pKa = 10.23EE310 pKa = 3.85VGSDD314 pKa = 3.38VEE316 pKa = 3.66ITYY319 pKa = 9.76IYY321 pKa = 10.16KK322 pKa = 9.86MKK324 pKa = 10.6VPKK327 pKa = 9.43TDD329 pKa = 3.07KK330 pKa = 10.98NLAAFLKK337 pKa = 10.3QVSAYY342 pKa = 9.25SQPSQAAEE350 pKa = 4.21VPSQLNPAAVVFQPLAEE367 pKa = 4.66DD368 pKa = 3.79EE369 pKa = 4.17QVEE372 pKa = 5.14IIDD375 pKa = 3.63QVYY378 pKa = 10.91GSFDD382 pKa = 3.02AA383 pKa = 5.67
MM1 pKa = 7.6ASVSFQEE8 pKa = 4.47EE9 pKa = 3.91KK10 pKa = 10.28RR11 pKa = 11.84GRR13 pKa = 11.84SGKK16 pKa = 10.09VPLSYY21 pKa = 10.71YY22 pKa = 10.67NPVIVSGDD30 pKa = 3.33KK31 pKa = 10.81PFWKK35 pKa = 10.34VMPNNAVPKK44 pKa = 10.6GKK46 pKa = 10.41GNKK49 pKa = 8.87DD50 pKa = 3.28QQIGYY55 pKa = 8.97WNEE58 pKa = 3.5QPRR61 pKa = 11.84YY62 pKa = 9.28RR63 pKa = 11.84MVRR66 pKa = 11.84GTRR69 pKa = 11.84KK70 pKa = 9.91DD71 pKa = 4.11LPSKK75 pKa = 8.43WHH77 pKa = 6.58FYY79 pKa = 10.71YY80 pKa = 10.91LGTGPHH86 pKa = 6.67AEE88 pKa = 3.88AKK90 pKa = 10.13FRR92 pKa = 11.84TRR94 pKa = 11.84TDD96 pKa = 2.94GVFWVAVQGSKK107 pKa = 9.39TEE109 pKa = 3.99PTGLGTRR116 pKa = 11.84KK117 pKa = 9.96RR118 pKa = 11.84NAEE121 pKa = 3.92LVNPEE126 pKa = 4.35FAIQLPAAIEE136 pKa = 4.03IQEE139 pKa = 4.22NTVSRR144 pKa = 11.84GNSRR148 pKa = 11.84NQSSNRR154 pKa = 11.84DD155 pKa = 3.21RR156 pKa = 11.84SQSGNRR162 pKa = 11.84SQNDD166 pKa = 3.31QKK168 pKa = 11.2QGNQNQKK175 pKa = 10.14SRR177 pKa = 11.84SNSQSRR183 pKa = 11.84KK184 pKa = 9.77GNQNSNDD191 pKa = 3.6SAVDD195 pKa = 2.96IVAAVKK201 pKa = 10.1QALKK205 pKa = 10.54EE206 pKa = 3.9LGVTNEE212 pKa = 4.09SNKK215 pKa = 9.98KK216 pKa = 9.45GKK218 pKa = 10.01NSGTTTPKK226 pKa = 9.52EE227 pKa = 4.05QRR229 pKa = 11.84SKK231 pKa = 11.36SPVRR235 pKa = 11.84SPTVQKK241 pKa = 10.58KK242 pKa = 7.53QLEE245 pKa = 4.23RR246 pKa = 11.84PPWKK250 pKa = 9.82RR251 pKa = 11.84VPNSTEE257 pKa = 4.38DD258 pKa = 3.29VTKK261 pKa = 10.8CFGVRR266 pKa = 11.84DD267 pKa = 3.94TYY269 pKa = 11.62RR270 pKa = 11.84NFGDD274 pKa = 3.78ADD276 pKa = 3.87LVRR279 pKa = 11.84NGIDD283 pKa = 3.11AKK285 pKa = 10.61HH286 pKa = 5.49YY287 pKa = 7.59PQLAEE292 pKa = 4.62LVPTPAAILFGGEE305 pKa = 4.17VVTKK309 pKa = 10.23EE310 pKa = 3.85VGSDD314 pKa = 3.38VEE316 pKa = 3.66ITYY319 pKa = 9.76IYY321 pKa = 10.16KK322 pKa = 9.86MKK324 pKa = 10.6VPKK327 pKa = 9.43TDD329 pKa = 3.07KK330 pKa = 10.98NLAAFLKK337 pKa = 10.3QVSAYY342 pKa = 9.25SQPSQAAEE350 pKa = 4.21VPSQLNPAAVVFQPLAEE367 pKa = 4.66DD368 pKa = 3.79EE369 pKa = 4.17QVEE372 pKa = 5.14IIDD375 pKa = 3.63QVYY378 pKa = 10.91GSFDD382 pKa = 3.02AA383 pKa = 5.67
Molecular weight: 42.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8899 |
65 |
6740 |
1483.2 |
165.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.54 ± 0.199 | 3.135 ± 0.805 |
5.113 ± 0.544 | 4.495 ± 0.269 |
5.652 ± 0.491 | 6.461 ± 0.46 |
1.697 ± 0.322 | 5.248 ± 0.639 |
6.046 ± 0.877 | 8.158 ± 0.681 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.068 ± 0.287 | 6.124 ± 0.453 |
3.528 ± 0.586 | 3.18 ± 0.95 |
3.225 ± 0.655 | 7.394 ± 0.5 |
5.967 ± 0.487 | 10.024 ± 0.324 |
1.18 ± 0.255 | 4.765 ± 0.306 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
