
Wenling sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

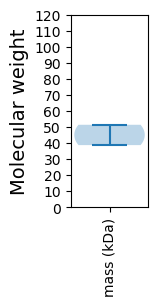
Protein with the lowest isoelectric point:
>tr|A0A1L3KF38|A0A1L3KF38_9VIRU RNA-directed RNA polymerase OS=Wenling sobemo-like virus 2 OX=1923541 PE=4 SV=1
MM1 pKa = 8.0EE2 pKa = 5.88IPDD5 pKa = 3.79DD6 pKa = 3.5WRR8 pKa = 11.84SDD10 pKa = 3.47VRR12 pKa = 11.84IDD14 pKa = 3.77QAIDD18 pKa = 3.34SLQGNSSPGVPFAHH32 pKa = 6.1VPSNNILRR40 pKa = 11.84EE41 pKa = 4.03KK42 pKa = 10.94YY43 pKa = 9.7RR44 pKa = 11.84GQLVTLVKK52 pKa = 10.41QRR54 pKa = 11.84LDD56 pKa = 3.47GNLPAFPIRR65 pKa = 11.84LFVKK69 pKa = 10.03PEE71 pKa = 3.28WHH73 pKa = 6.94KK74 pKa = 10.79IEE76 pKa = 4.78KK77 pKa = 9.88LRR79 pKa = 11.84QQRR82 pKa = 11.84YY83 pKa = 8.37RR84 pKa = 11.84LISQVSVVDD93 pKa = 3.54QVIDD97 pKa = 3.61KK98 pKa = 10.75LLVASYY104 pKa = 10.87YY105 pKa = 10.68DD106 pKa = 3.91YY107 pKa = 11.61EE108 pKa = 4.13NSHH111 pKa = 7.07YY112 pKa = 10.13IRR114 pKa = 11.84QPGKK118 pKa = 10.64VGWSYY123 pKa = 11.89VGGGYY128 pKa = 10.71KK129 pKa = 10.29LVPRR133 pKa = 11.84GWGYY137 pKa = 10.82DD138 pKa = 3.1RR139 pKa = 11.84KK140 pKa = 10.33AWDD143 pKa = 3.22WSVPSWLLEE152 pKa = 4.29DD153 pKa = 4.14YY154 pKa = 11.26EE155 pKa = 5.19FFLEE159 pKa = 4.23NQITSTKK166 pKa = 9.63DD167 pKa = 2.86HH168 pKa = 7.03KK169 pKa = 11.07DD170 pKa = 2.85WLRR173 pKa = 11.84KK174 pKa = 9.48RR175 pKa = 11.84FAEE178 pKa = 4.5LYY180 pKa = 8.41EE181 pKa = 4.37RR182 pKa = 11.84PILQLSDD189 pKa = 3.26GTLLQQLVSGVQKK202 pKa = 10.71SGAYY206 pKa = 7.44YY207 pKa = 9.59TLNGNTTMQVLLHH220 pKa = 6.53HH221 pKa = 6.75ATTLEE226 pKa = 4.1SGEE229 pKa = 5.05DD230 pKa = 3.87STPMIALGDD239 pKa = 3.89DD240 pKa = 3.81TVQPLASPAYY250 pKa = 10.62LEE252 pKa = 4.23VLQRR256 pKa = 11.84WVTLKK261 pKa = 10.56EE262 pKa = 3.73AVPNEE267 pKa = 4.2FCGMSYY273 pKa = 10.22SGEE276 pKa = 4.08NVEE279 pKa = 5.47PVYY282 pKa = 10.52QDD284 pKa = 3.01KK285 pKa = 10.95HH286 pKa = 7.69AMNLAADD293 pKa = 4.77LSQDD297 pKa = 3.2TLLQYY302 pKa = 10.96HH303 pKa = 7.13LLYY306 pKa = 10.6GQSTKK311 pKa = 10.66LPKK314 pKa = 10.04LRR316 pKa = 11.84RR317 pKa = 11.84LNALAGHH324 pKa = 6.67PPVHH328 pKa = 5.94QLWLSTIWGG337 pKa = 3.72
MM1 pKa = 8.0EE2 pKa = 5.88IPDD5 pKa = 3.79DD6 pKa = 3.5WRR8 pKa = 11.84SDD10 pKa = 3.47VRR12 pKa = 11.84IDD14 pKa = 3.77QAIDD18 pKa = 3.34SLQGNSSPGVPFAHH32 pKa = 6.1VPSNNILRR40 pKa = 11.84EE41 pKa = 4.03KK42 pKa = 10.94YY43 pKa = 9.7RR44 pKa = 11.84GQLVTLVKK52 pKa = 10.41QRR54 pKa = 11.84LDD56 pKa = 3.47GNLPAFPIRR65 pKa = 11.84LFVKK69 pKa = 10.03PEE71 pKa = 3.28WHH73 pKa = 6.94KK74 pKa = 10.79IEE76 pKa = 4.78KK77 pKa = 9.88LRR79 pKa = 11.84QQRR82 pKa = 11.84YY83 pKa = 8.37RR84 pKa = 11.84LISQVSVVDD93 pKa = 3.54QVIDD97 pKa = 3.61KK98 pKa = 10.75LLVASYY104 pKa = 10.87YY105 pKa = 10.68DD106 pKa = 3.91YY107 pKa = 11.61EE108 pKa = 4.13NSHH111 pKa = 7.07YY112 pKa = 10.13IRR114 pKa = 11.84QPGKK118 pKa = 10.64VGWSYY123 pKa = 11.89VGGGYY128 pKa = 10.71KK129 pKa = 10.29LVPRR133 pKa = 11.84GWGYY137 pKa = 10.82DD138 pKa = 3.1RR139 pKa = 11.84KK140 pKa = 10.33AWDD143 pKa = 3.22WSVPSWLLEE152 pKa = 4.29DD153 pKa = 4.14YY154 pKa = 11.26EE155 pKa = 5.19FFLEE159 pKa = 4.23NQITSTKK166 pKa = 9.63DD167 pKa = 2.86HH168 pKa = 7.03KK169 pKa = 11.07DD170 pKa = 2.85WLRR173 pKa = 11.84KK174 pKa = 9.48RR175 pKa = 11.84FAEE178 pKa = 4.5LYY180 pKa = 8.41EE181 pKa = 4.37RR182 pKa = 11.84PILQLSDD189 pKa = 3.26GTLLQQLVSGVQKK202 pKa = 10.71SGAYY206 pKa = 7.44YY207 pKa = 9.59TLNGNTTMQVLLHH220 pKa = 6.53HH221 pKa = 6.75ATTLEE226 pKa = 4.1SGEE229 pKa = 5.05DD230 pKa = 3.87STPMIALGDD239 pKa = 3.89DD240 pKa = 3.81TVQPLASPAYY250 pKa = 10.62LEE252 pKa = 4.23VLQRR256 pKa = 11.84WVTLKK261 pKa = 10.56EE262 pKa = 3.73AVPNEE267 pKa = 4.2FCGMSYY273 pKa = 10.22SGEE276 pKa = 4.08NVEE279 pKa = 5.47PVYY282 pKa = 10.52QDD284 pKa = 3.01KK285 pKa = 10.95HH286 pKa = 7.69AMNLAADD293 pKa = 4.77LSQDD297 pKa = 3.2TLLQYY302 pKa = 10.96HH303 pKa = 7.13LLYY306 pKa = 10.6GQSTKK311 pKa = 10.66LPKK314 pKa = 10.04LRR316 pKa = 11.84RR317 pKa = 11.84LNALAGHH324 pKa = 6.67PPVHH328 pKa = 5.94QLWLSTIWGG337 pKa = 3.72
Molecular weight: 38.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF38|A0A1L3KF38_9VIRU RNA-directed RNA polymerase OS=Wenling sobemo-like virus 2 OX=1923541 PE=4 SV=1
MM1 pKa = 7.32FKK3 pKa = 10.61VIAIGVVLATLVALARR19 pKa = 11.84YY20 pKa = 8.7RR21 pKa = 11.84WDD23 pKa = 3.38YY24 pKa = 10.87LVTMGALAMVFVMLVWEE41 pKa = 4.14LLFRR45 pKa = 11.84FWQGALVPLYY55 pKa = 10.45EE56 pKa = 4.59SLLEE60 pKa = 4.03PLLVGDD66 pKa = 4.79DD67 pKa = 4.04PTPIFYY73 pKa = 9.67TPTPTTMEE81 pKa = 4.56LTPMMIYY88 pKa = 10.47FMIVAATVILVTLIVRR104 pKa = 11.84CTPSSRR110 pKa = 11.84IIYY113 pKa = 9.73SPEE116 pKa = 3.62AMVHH120 pKa = 5.97GSAFEE125 pKa = 3.96ALPPPKK131 pKa = 10.43CQVTLVAVTGTEE143 pKa = 3.9VSTGQGALIKK153 pKa = 10.1YY154 pKa = 8.98GEE156 pKa = 4.54GVAVVTALHH165 pKa = 5.38VVKK168 pKa = 10.42GASVVLAEE176 pKa = 4.09NRR178 pKa = 11.84EE179 pKa = 4.09GLSIRR184 pKa = 11.84LGARR188 pKa = 11.84KK189 pKa = 9.64VIHH192 pKa = 6.18QDD194 pKa = 2.85GVAFPVEE201 pKa = 3.99AAQMSRR207 pKa = 11.84LGVRR211 pKa = 11.84VGQTTVVTSPVAASAHH227 pKa = 4.65GVMGRR232 pKa = 11.84SSGCLMQISDD242 pKa = 4.87GLLSYY247 pKa = 10.52QGSTQPGYY255 pKa = 10.55SGAPVMVGGKK265 pKa = 9.35VAGIHH270 pKa = 6.16IGTVEE275 pKa = 3.9GRR277 pKa = 11.84NLAFDD282 pKa = 3.54ISLAISRR289 pKa = 11.84SNDD292 pKa = 2.92YY293 pKa = 10.42RR294 pKa = 11.84YY295 pKa = 10.62NPEE298 pKa = 3.82AQTTSEE304 pKa = 3.85MTPRR308 pKa = 11.84EE309 pKa = 3.69KK310 pKa = 10.61GYY312 pKa = 10.78YY313 pKa = 10.13GGMDD317 pKa = 3.12ARR319 pKa = 11.84FKK321 pKa = 11.01KK322 pKa = 10.53GLAKK326 pKa = 10.61LEE328 pKa = 4.28EE329 pKa = 4.38QDD331 pKa = 4.3DD332 pKa = 4.32QPGSWAHH339 pKa = 5.95IPEE342 pKa = 4.84EE343 pKa = 3.8IDD345 pKa = 3.3YY346 pKa = 10.98SRR348 pKa = 11.84KK349 pKa = 8.54LTFEE353 pKa = 3.91NQMLSLTKK361 pKa = 10.35SVAALEE367 pKa = 4.17EE368 pKa = 4.55TVAKK372 pKa = 9.8MDD374 pKa = 3.41FRR376 pKa = 11.84RR377 pKa = 11.84RR378 pKa = 11.84RR379 pKa = 11.84GSTRR383 pKa = 11.84RR384 pKa = 11.84HH385 pKa = 5.51LSSSSEE391 pKa = 4.23SSCSSSEE398 pKa = 3.9SSRR401 pKa = 11.84DD402 pKa = 2.98EE403 pKa = 4.19DD404 pKa = 3.48RR405 pKa = 11.84GSRR408 pKa = 11.84PSQKK412 pKa = 10.51SKK414 pKa = 8.64NHH416 pKa = 6.3RR417 pKa = 11.84KK418 pKa = 9.63RR419 pKa = 11.84PSKK422 pKa = 10.2HH423 pKa = 5.85LRR425 pKa = 11.84RR426 pKa = 11.84QASRR430 pKa = 11.84ISRR433 pKa = 11.84KK434 pKa = 9.66SVVVTVPHH442 pKa = 6.88PSEE445 pKa = 4.77PPPKK449 pKa = 9.34PGPSGLTSSTSGKK462 pKa = 10.54GPAHH466 pKa = 6.45PKK468 pKa = 10.06KK469 pKa = 10.85SEE471 pKa = 3.8GKK473 pKa = 9.0QQ474 pKa = 3.09
MM1 pKa = 7.32FKK3 pKa = 10.61VIAIGVVLATLVALARR19 pKa = 11.84YY20 pKa = 8.7RR21 pKa = 11.84WDD23 pKa = 3.38YY24 pKa = 10.87LVTMGALAMVFVMLVWEE41 pKa = 4.14LLFRR45 pKa = 11.84FWQGALVPLYY55 pKa = 10.45EE56 pKa = 4.59SLLEE60 pKa = 4.03PLLVGDD66 pKa = 4.79DD67 pKa = 4.04PTPIFYY73 pKa = 9.67TPTPTTMEE81 pKa = 4.56LTPMMIYY88 pKa = 10.47FMIVAATVILVTLIVRR104 pKa = 11.84CTPSSRR110 pKa = 11.84IIYY113 pKa = 9.73SPEE116 pKa = 3.62AMVHH120 pKa = 5.97GSAFEE125 pKa = 3.96ALPPPKK131 pKa = 10.43CQVTLVAVTGTEE143 pKa = 3.9VSTGQGALIKK153 pKa = 10.1YY154 pKa = 8.98GEE156 pKa = 4.54GVAVVTALHH165 pKa = 5.38VVKK168 pKa = 10.42GASVVLAEE176 pKa = 4.09NRR178 pKa = 11.84EE179 pKa = 4.09GLSIRR184 pKa = 11.84LGARR188 pKa = 11.84KK189 pKa = 9.64VIHH192 pKa = 6.18QDD194 pKa = 2.85GVAFPVEE201 pKa = 3.99AAQMSRR207 pKa = 11.84LGVRR211 pKa = 11.84VGQTTVVTSPVAASAHH227 pKa = 4.65GVMGRR232 pKa = 11.84SSGCLMQISDD242 pKa = 4.87GLLSYY247 pKa = 10.52QGSTQPGYY255 pKa = 10.55SGAPVMVGGKK265 pKa = 9.35VAGIHH270 pKa = 6.16IGTVEE275 pKa = 3.9GRR277 pKa = 11.84NLAFDD282 pKa = 3.54ISLAISRR289 pKa = 11.84SNDD292 pKa = 2.92YY293 pKa = 10.42RR294 pKa = 11.84YY295 pKa = 10.62NPEE298 pKa = 3.82AQTTSEE304 pKa = 3.85MTPRR308 pKa = 11.84EE309 pKa = 3.69KK310 pKa = 10.61GYY312 pKa = 10.78YY313 pKa = 10.13GGMDD317 pKa = 3.12ARR319 pKa = 11.84FKK321 pKa = 11.01KK322 pKa = 10.53GLAKK326 pKa = 10.61LEE328 pKa = 4.28EE329 pKa = 4.38QDD331 pKa = 4.3DD332 pKa = 4.32QPGSWAHH339 pKa = 5.95IPEE342 pKa = 4.84EE343 pKa = 3.8IDD345 pKa = 3.3YY346 pKa = 10.98SRR348 pKa = 11.84KK349 pKa = 8.54LTFEE353 pKa = 3.91NQMLSLTKK361 pKa = 10.35SVAALEE367 pKa = 4.17EE368 pKa = 4.55TVAKK372 pKa = 9.8MDD374 pKa = 3.41FRR376 pKa = 11.84RR377 pKa = 11.84RR378 pKa = 11.84RR379 pKa = 11.84GSTRR383 pKa = 11.84RR384 pKa = 11.84HH385 pKa = 5.51LSSSSEE391 pKa = 4.23SSCSSSEE398 pKa = 3.9SSRR401 pKa = 11.84DD402 pKa = 2.98EE403 pKa = 4.19DD404 pKa = 3.48RR405 pKa = 11.84GSRR408 pKa = 11.84PSQKK412 pKa = 10.51SKK414 pKa = 8.64NHH416 pKa = 6.3RR417 pKa = 11.84KK418 pKa = 9.63RR419 pKa = 11.84PSKK422 pKa = 10.2HH423 pKa = 5.85LRR425 pKa = 11.84RR426 pKa = 11.84QASRR430 pKa = 11.84ISRR433 pKa = 11.84KK434 pKa = 9.66SVVVTVPHH442 pKa = 6.88PSEE445 pKa = 4.77PPPKK449 pKa = 9.34PGPSGLTSSTSGKK462 pKa = 10.54GPAHH466 pKa = 6.45PKK468 pKa = 10.06KK469 pKa = 10.85SEE471 pKa = 3.8GKK473 pKa = 9.0QQ474 pKa = 3.09
Molecular weight: 51.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
811 |
337 |
474 |
405.5 |
45.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.782 ± 1.101 | 0.617 ± 0.203 |
4.316 ± 1.214 | 5.425 ± 0.241 |
2.343 ± 0.168 | 7.645 ± 0.708 |
2.589 ± 0.24 | 4.069 ± 0.134 |
4.932 ± 0.071 | 10.111 ± 1.679 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.713 ± 0.779 | 2.219 ± 0.85 |
6.165 ± 0.334 | 4.686 ± 1.168 |
6.165 ± 0.522 | 9.125 ± 1.269 |
5.672 ± 0.774 | 8.631 ± 0.769 |
1.85 ± 0.896 | 3.946 ± 0.884 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
