
Penicillium aurantiogriseum bipartite virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
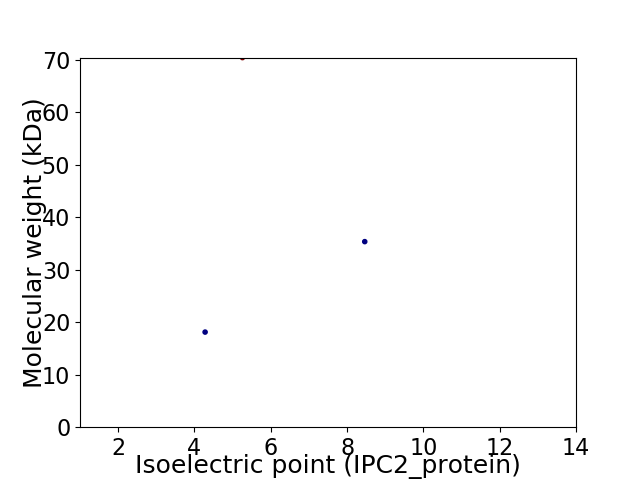
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2KPE1|A0A0S2KPE1_9VIRU 20 kDa protein OS=Penicillium aurantiogriseum bipartite virus 1 OX=1755754 PE=4 SV=1
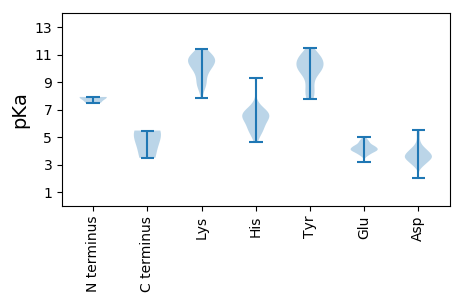
MM1 pKa = 7.49VGVGSDD7 pKa = 3.01TDD9 pKa = 4.04GNDD12 pKa = 2.95EE13 pKa = 4.34HH14 pKa = 9.29SYY16 pKa = 8.8EE17 pKa = 4.05QWRR20 pKa = 11.84TTPLDD25 pKa = 3.58VIIARR30 pKa = 11.84DD31 pKa = 3.42WVVEE35 pKa = 3.88LEE37 pKa = 4.09QGPMEE42 pKa = 4.81LMAEE46 pKa = 4.38LGLRR50 pKa = 11.84SPSYY54 pKa = 10.66PDD56 pKa = 4.19LEE58 pKa = 4.7HH59 pKa = 6.55HH60 pKa = 6.5ATWISDD66 pKa = 3.59DD67 pKa = 4.63DD68 pKa = 5.29GDD70 pKa = 3.92VTRR73 pKa = 11.84TGVPYY78 pKa = 10.49RR79 pKa = 11.84GEE81 pKa = 3.78NLFAQWVLEE90 pKa = 4.03TLLGEE95 pKa = 4.18SRR97 pKa = 11.84MDD99 pKa = 3.37VEE101 pKa = 4.51LTAEE105 pKa = 4.31VAAEE109 pKa = 4.01MGVMAVAGYY118 pKa = 9.7SAAVWLQGEE127 pKa = 4.58VIDD130 pKa = 5.46LRR132 pKa = 11.84VLRR135 pKa = 11.84VASTPLAQAYY145 pKa = 7.8HH146 pKa = 6.63ALVAGKK152 pKa = 10.0SAVDD156 pKa = 3.59EE157 pKa = 4.19KK158 pKa = 11.37HH159 pKa = 6.66FMGGSANN166 pKa = 3.49
MM1 pKa = 7.49VGVGSDD7 pKa = 3.01TDD9 pKa = 4.04GNDD12 pKa = 2.95EE13 pKa = 4.34HH14 pKa = 9.29SYY16 pKa = 8.8EE17 pKa = 4.05QWRR20 pKa = 11.84TTPLDD25 pKa = 3.58VIIARR30 pKa = 11.84DD31 pKa = 3.42WVVEE35 pKa = 3.88LEE37 pKa = 4.09QGPMEE42 pKa = 4.81LMAEE46 pKa = 4.38LGLRR50 pKa = 11.84SPSYY54 pKa = 10.66PDD56 pKa = 4.19LEE58 pKa = 4.7HH59 pKa = 6.55HH60 pKa = 6.5ATWISDD66 pKa = 3.59DD67 pKa = 4.63DD68 pKa = 5.29GDD70 pKa = 3.92VTRR73 pKa = 11.84TGVPYY78 pKa = 10.49RR79 pKa = 11.84GEE81 pKa = 3.78NLFAQWVLEE90 pKa = 4.03TLLGEE95 pKa = 4.18SRR97 pKa = 11.84MDD99 pKa = 3.37VEE101 pKa = 4.51LTAEE105 pKa = 4.31VAAEE109 pKa = 4.01MGVMAVAGYY118 pKa = 9.7SAAVWLQGEE127 pKa = 4.58VIDD130 pKa = 5.46LRR132 pKa = 11.84VLRR135 pKa = 11.84VASTPLAQAYY145 pKa = 7.8HH146 pKa = 6.63ALVAGKK152 pKa = 10.0SAVDD156 pKa = 3.59EE157 pKa = 4.19KK158 pKa = 11.37HH159 pKa = 6.66FMGGSANN166 pKa = 3.49
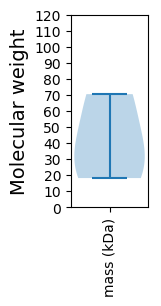
Molecular weight: 18.13 kDa
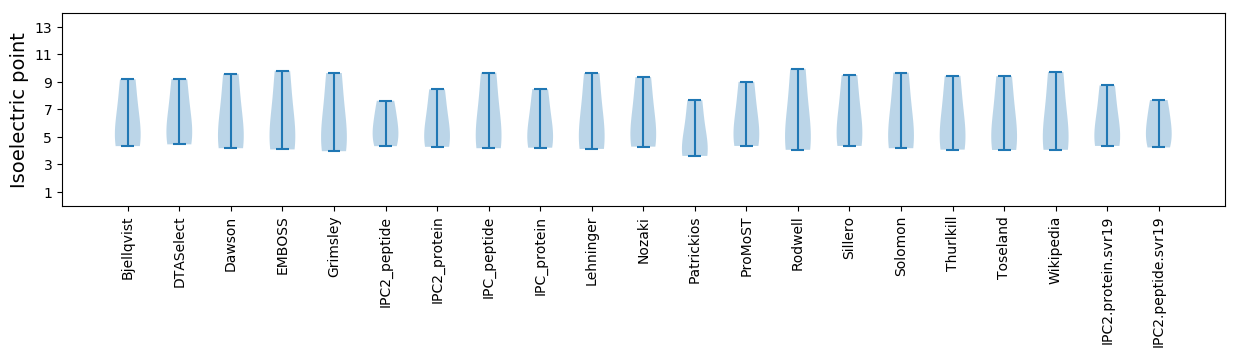
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2KPE1|A0A0S2KPE1_9VIRU 20 kDa protein OS=Penicillium aurantiogriseum bipartite virus 1 OX=1755754 PE=4 SV=1
MM1 pKa = 7.92ADD3 pKa = 3.1MTAPPSFLDD12 pKa = 3.4KK13 pKa = 11.12FGGSTANVDD22 pKa = 3.74YY23 pKa = 10.96TDD25 pKa = 5.13DD26 pKa = 3.92PSKK29 pKa = 10.92QGGILPIPLQGDD41 pKa = 3.68SLATLEE47 pKa = 4.31TKK49 pKa = 9.22MRR51 pKa = 11.84NLARR55 pKa = 11.84VVEE58 pKa = 4.99RR59 pKa = 11.84GASVARR65 pKa = 11.84FTGMMGTPQQSRR77 pKa = 11.84DD78 pKa = 3.41ALQTAKK84 pKa = 10.02MRR86 pKa = 11.84MTEE89 pKa = 3.93VEE91 pKa = 4.4KK92 pKa = 11.03EE93 pKa = 4.21MFSHH97 pKa = 6.56WEE99 pKa = 3.68AGGRR103 pKa = 11.84LPDD106 pKa = 4.03VNWGTDD112 pKa = 3.18VDD114 pKa = 4.18AMPSGSEE121 pKa = 3.84PRR123 pKa = 11.84RR124 pKa = 11.84KK125 pKa = 10.23AEE127 pKa = 3.9IQAEE131 pKa = 4.11ALNRR135 pKa = 11.84LYY137 pKa = 9.97KK138 pKa = 10.14TDD140 pKa = 3.49AATAGHH146 pKa = 5.35VVHH149 pKa = 7.02WKK151 pKa = 7.86KK152 pKa = 8.86TFRR155 pKa = 11.84AIIPLLVSMTKK166 pKa = 10.1VVGSEE171 pKa = 3.7RR172 pKa = 11.84DD173 pKa = 3.4FLGGQDD179 pKa = 3.52VLDD182 pKa = 4.5ANEE185 pKa = 3.93MAEE188 pKa = 4.02VQTIGKK194 pKa = 9.73VNGTTYY200 pKa = 11.44ASINAIQEE208 pKa = 4.06EE209 pKa = 4.64VHH211 pKa = 6.65RR212 pKa = 11.84LLSSINRR219 pKa = 11.84SKK221 pKa = 11.43DD222 pKa = 3.17LLQARR227 pKa = 11.84EE228 pKa = 4.02HH229 pKa = 6.31ALKK232 pKa = 10.75AKK234 pKa = 9.53NPKK237 pKa = 10.08YY238 pKa = 10.12KK239 pKa = 10.52VKK241 pKa = 10.63QSPANDD247 pKa = 2.9ARR249 pKa = 11.84QLLGKK254 pKa = 9.82RR255 pKa = 11.84RR256 pKa = 11.84LGADD260 pKa = 3.03VDD262 pKa = 5.14FSADD266 pKa = 3.91PIPEE270 pKa = 3.6RR271 pKa = 11.84RR272 pKa = 11.84VRR274 pKa = 11.84RR275 pKa = 11.84NLALGVHH282 pKa = 5.39QEE284 pKa = 4.26SVPIASRR291 pKa = 11.84AGEE294 pKa = 3.83QRR296 pKa = 11.84APTRR300 pKa = 11.84GGINQRR306 pKa = 11.84RR307 pKa = 11.84PHH309 pKa = 7.31PDD311 pKa = 3.13TDD313 pKa = 3.77PSADD317 pKa = 3.54MDD319 pKa = 4.02TQII322 pKa = 5.11
MM1 pKa = 7.92ADD3 pKa = 3.1MTAPPSFLDD12 pKa = 3.4KK13 pKa = 11.12FGGSTANVDD22 pKa = 3.74YY23 pKa = 10.96TDD25 pKa = 5.13DD26 pKa = 3.92PSKK29 pKa = 10.92QGGILPIPLQGDD41 pKa = 3.68SLATLEE47 pKa = 4.31TKK49 pKa = 9.22MRR51 pKa = 11.84NLARR55 pKa = 11.84VVEE58 pKa = 4.99RR59 pKa = 11.84GASVARR65 pKa = 11.84FTGMMGTPQQSRR77 pKa = 11.84DD78 pKa = 3.41ALQTAKK84 pKa = 10.02MRR86 pKa = 11.84MTEE89 pKa = 3.93VEE91 pKa = 4.4KK92 pKa = 11.03EE93 pKa = 4.21MFSHH97 pKa = 6.56WEE99 pKa = 3.68AGGRR103 pKa = 11.84LPDD106 pKa = 4.03VNWGTDD112 pKa = 3.18VDD114 pKa = 4.18AMPSGSEE121 pKa = 3.84PRR123 pKa = 11.84RR124 pKa = 11.84KK125 pKa = 10.23AEE127 pKa = 3.9IQAEE131 pKa = 4.11ALNRR135 pKa = 11.84LYY137 pKa = 9.97KK138 pKa = 10.14TDD140 pKa = 3.49AATAGHH146 pKa = 5.35VVHH149 pKa = 7.02WKK151 pKa = 7.86KK152 pKa = 8.86TFRR155 pKa = 11.84AIIPLLVSMTKK166 pKa = 10.1VVGSEE171 pKa = 3.7RR172 pKa = 11.84DD173 pKa = 3.4FLGGQDD179 pKa = 3.52VLDD182 pKa = 4.5ANEE185 pKa = 3.93MAEE188 pKa = 4.02VQTIGKK194 pKa = 9.73VNGTTYY200 pKa = 11.44ASINAIQEE208 pKa = 4.06EE209 pKa = 4.64VHH211 pKa = 6.65RR212 pKa = 11.84LLSSINRR219 pKa = 11.84SKK221 pKa = 11.43DD222 pKa = 3.17LLQARR227 pKa = 11.84EE228 pKa = 4.02HH229 pKa = 6.31ALKK232 pKa = 10.75AKK234 pKa = 9.53NPKK237 pKa = 10.08YY238 pKa = 10.12KK239 pKa = 10.52VKK241 pKa = 10.63QSPANDD247 pKa = 2.9ARR249 pKa = 11.84QLLGKK254 pKa = 9.82RR255 pKa = 11.84RR256 pKa = 11.84LGADD260 pKa = 3.03VDD262 pKa = 5.14FSADD266 pKa = 3.91PIPEE270 pKa = 3.6RR271 pKa = 11.84RR272 pKa = 11.84VRR274 pKa = 11.84RR275 pKa = 11.84NLALGVHH282 pKa = 5.39QEE284 pKa = 4.26SVPIASRR291 pKa = 11.84AGEE294 pKa = 3.83QRR296 pKa = 11.84APTRR300 pKa = 11.84GGINQRR306 pKa = 11.84RR307 pKa = 11.84PHH309 pKa = 7.31PDD311 pKa = 3.13TDD313 pKa = 3.77PSADD317 pKa = 3.54MDD319 pKa = 4.02TQII322 pKa = 5.11
Molecular weight: 35.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1102 |
166 |
614 |
367.3 |
41.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.53 ± 1.296 | 0.454 ± 0.276 |
7.26 ± 0.149 | 7.169 ± 0.686 |
3.085 ± 0.78 | 6.897 ± 0.735 |
2.269 ± 0.197 | 4.446 ± 0.666 |
4.537 ± 0.869 | 8.53 ± 0.453 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.722 ± 0.72 | 2.904 ± 0.372 |
5.626 ± 0.531 | 4.628 ± 0.42 |
7.078 ± 0.669 | 5.626 ± 0.319 |
5.898 ± 0.15 | 7.169 ± 0.96 |
1.996 ± 0.442 | 3.176 ± 0.869 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
