
Candidatus Rhodobacter lobularis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodobacter
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
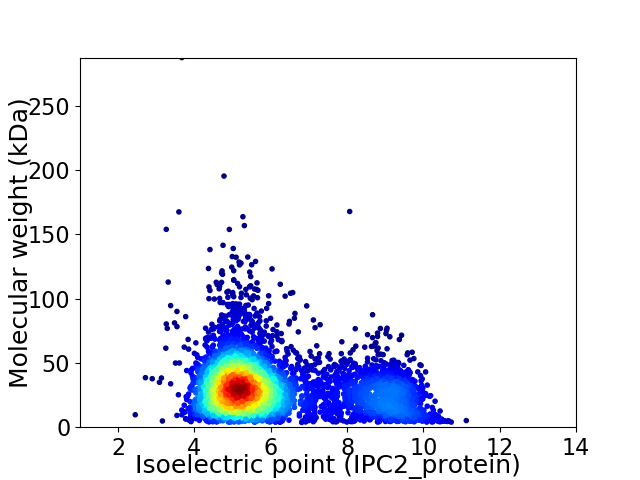
Virtual 2D-PAGE plot for 4763 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
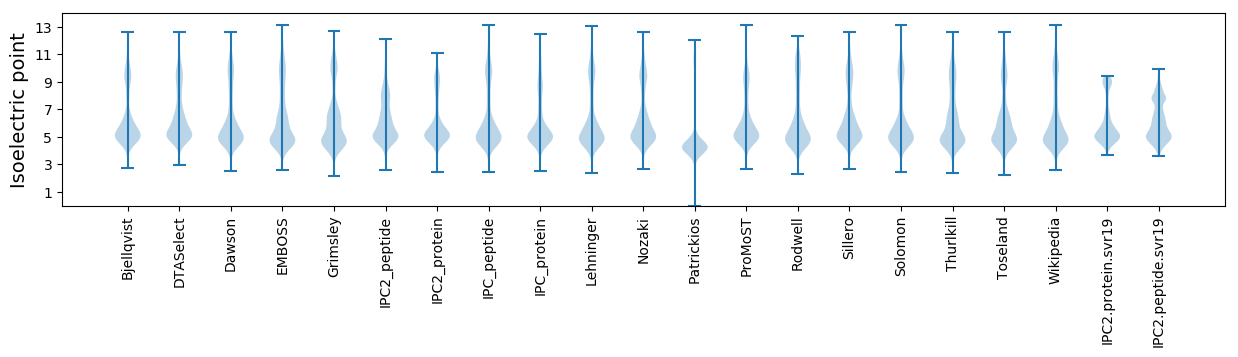
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0J9E1N7|A0A0J9E1N7_9RHOB Sarcosine oxidase beta subunit OS=Candidatus Rhodobacter lobularis OX=1675527 GN=AIOL_001767 PE=4 SV=1
MM1 pKa = 7.82SIFSKK6 pKa = 10.83LNRR9 pKa = 11.84ALSAIFFVAVAAAGSPSWAQPAFTVGFTPNSIAPSGTTRR48 pKa = 11.84MIFTIDD54 pKa = 3.46NSSGGAVTSMDD65 pKa = 3.65FTNALPAGITVAVAPNAFTSCGGASASSATVTAMAGGSSIAMADD109 pKa = 3.62GQLAPGATCTVEE121 pKa = 4.45ANVTATATATNGPATLNTSNGSSASASVGLTVDD154 pKa = 3.8AAATAVGKK162 pKa = 10.54AFAPTTIGIGGVSRR176 pKa = 11.84LTLTVDD182 pKa = 3.55NSAGANAQNFTLTDD196 pKa = 3.64NLPAGMVLADD206 pKa = 4.47PPNTATTCPVAQSQTLTGVGGGSILEE232 pKa = 4.16LQSTGGLVFTPSFMVVNALPAGTVCTVSADD262 pKa = 3.55VTASVPGSLQNVTEE276 pKa = 4.34PFLDD280 pKa = 3.91SGSAAATLTVTGAPAGAPQLTKK302 pKa = 10.77EE303 pKa = 4.24FTDD306 pKa = 4.14DD307 pKa = 3.28PVAAGGSATLEE318 pKa = 3.78FEE320 pKa = 3.78ILNRR324 pKa = 11.84SRR326 pKa = 11.84SDD328 pKa = 3.08AASNIAFTDD337 pKa = 3.53DD338 pKa = 2.98MSALFGALAGTVTAIGTNTCGGTPSGVGTSSLSYY372 pKa = 10.75SGGTVGAGLRR382 pKa = 11.84CNITVTISVPAGVSTGSYY400 pKa = 9.24TNTSTAVTADD410 pKa = 3.48VGGMSLTGNAATANLSVLAALDD432 pKa = 4.11LPPVLSFTTPDD443 pKa = 3.23GNAGGVSTWSFTLSNAASSALTDD466 pKa = 3.42GTFDD470 pKa = 3.49VQMNPPLPFPTNASLPPMPDD490 pKa = 3.29PPCGVGSSLSLAFLDD505 pKa = 4.1TDD507 pKa = 3.68QQALRR512 pKa = 11.84LTGASLATSGAAGDD526 pKa = 3.88SCTFDD531 pKa = 3.27VDD533 pKa = 3.37IGLNSDD539 pKa = 5.14LPPLQYY545 pKa = 11.04NFTSGAPSGTVNGATRR561 pKa = 11.84TGTPASSSMTVGTNLNLSFAKK582 pKa = 10.45SFLATAVPGGTVDD595 pKa = 3.94LEE597 pKa = 4.28FVITSSGDD605 pKa = 3.46STDD608 pKa = 2.91ATAISFTDD616 pKa = 5.0DD617 pKa = 3.09INAMLAGTSLNAVISNEE634 pKa = 4.01CGGNLTGTTNLSYY647 pKa = 10.57TGGSLTGGTSCAIVTRR663 pKa = 11.84LDD665 pKa = 3.43VPAAAAIGTYY675 pKa = 10.32TNTTSALSATPSGSSTVEE693 pKa = 3.91TFGTASADD701 pKa = 3.47LTVAGLTFEE710 pKa = 5.78KK711 pKa = 10.68EE712 pKa = 4.04FLTNTVLAGDD722 pKa = 3.84TTTLRR727 pKa = 11.84FTISNVHH734 pKa = 5.72PTADD738 pKa = 3.23ATITSFTDD746 pKa = 3.15NLQQNLTGLAATGGPSGNTCGGTLSGTTFLIYY778 pKa = 10.26TGGSVTSGMSCTIDD792 pKa = 3.52VEE794 pKa = 4.53VLVPVGTADD803 pKa = 3.25GSYY806 pKa = 11.42GNTTSTLSATQGGGPVVVDD825 pKa = 4.0PATDD829 pKa = 3.14ILNVQGLQIEE839 pKa = 4.43LSKK842 pKa = 11.32AFTDD846 pKa = 4.42DD847 pKa = 3.45PVAPGDD853 pKa = 3.64NATLEE858 pKa = 4.41FTLSNLSSTEE868 pKa = 4.12AISNIAFTDD877 pKa = 3.5DD878 pKa = 3.76LAAMLPGTTFASVAGNDD895 pKa = 3.41CGATVGGTGTSTISVSGTSLAASATCTITVAVTVPGGAVDD935 pKa = 3.52NTYY938 pKa = 10.96TNTTSSVTGDD948 pKa = 2.79IGGLAVTGDD957 pKa = 3.84PATAQLSVSSILPLAFSKK975 pKa = 10.94SFSGSVASGEE985 pKa = 4.37SGTLTFTLSNPNTTALSDD1003 pKa = 4.2LGFTDD1008 pKa = 5.29DD1009 pKa = 4.78LNAMLTGVVATSLPSAPCGLGSSITGTSSLTASGFSLDD1047 pKa = 4.27ASGGSCTFDD1056 pKa = 2.78VGFSVPVTATGATYY1070 pKa = 11.21LNTTSEE1076 pKa = 4.37LVSFGLSLAPPATANLIVTDD1096 pKa = 4.52PVADD1100 pKa = 4.37LSLAIADD1107 pKa = 4.24SPDD1110 pKa = 3.59PVTPGQNLVYY1120 pKa = 10.12TVTASNAGPQTAQSVAVNLPVPTGTTLVSTTGCAEE1155 pKa = 4.41DD1156 pKa = 3.86PSGAATCSLGNIASGGNAQYY1176 pKa = 10.53TLTVTVDD1183 pKa = 3.0AATSGTVSASGTVSSSTSDD1202 pKa = 3.19PVAGNNTATASTSAPATADD1221 pKa = 3.12ISITKK1226 pKa = 10.16SDD1228 pKa = 3.63GVTSANSGGTTTYY1241 pKa = 10.78SIVVANAGPSADD1253 pKa = 3.74PAVSVADD1260 pKa = 3.92TFAADD1265 pKa = 4.66LGCATTSVAAGGATGNTAAFSGDD1288 pKa = 3.07IAEE1291 pKa = 4.46TLSMPASSSVTYY1303 pKa = 9.09TATCDD1308 pKa = 3.17IAITATGTLSNTATATASVLDD1329 pKa = 4.37PNATNNSATDD1339 pKa = 3.17SDD1341 pKa = 4.34TALVAPVAMGFSKK1354 pKa = 10.76AFSPATIDD1362 pKa = 3.19QGANTTLTFTIDD1374 pKa = 3.04NTANAVAADD1383 pKa = 3.92NLNFTDD1389 pKa = 5.15GFPLGVDD1396 pKa = 3.92LSVAAPVVSSSTCGGTFNPVAGGGNLVFSGGSVAAGATCTVSVLLNARR1444 pKa = 11.84GSGTATNTSSQLGSFFPTPSAAATADD1470 pKa = 3.23ITVNPAVAPTFAKK1483 pKa = 10.65AFGAATIEE1491 pKa = 4.06QGQTTTATFTINNSANSIEE1510 pKa = 4.14ASSLQFSDD1518 pKa = 3.71TLPVEE1523 pKa = 4.44LVVAATPNASTTCTGGTITATAGSGSILYY1552 pKa = 10.56SGGEE1556 pKa = 4.03VAAGATCTVTVTVDD1570 pKa = 3.32SLGSGSIANPSLNLTSSLPTATAAATPLTINPATAPGVAKK1610 pKa = 9.81TFTPSAITQGEE1621 pKa = 4.31TSALSITIDD1630 pKa = 3.12NSANFLEE1637 pKa = 4.45MTGVAFTDD1645 pKa = 4.08AFPSGMVVGPTPGASNTCGGTFSASAGSGTISLSGGTVAAGATCSVGVTVRR1696 pKa = 11.84GNAAGTLTNPAFDD1709 pKa = 3.67VTSNIATGTAAATDD1723 pKa = 4.08LAVNPAAAPGFAKK1736 pKa = 10.65AFGAGTINQGGTVVATVSVDD1756 pKa = 3.39NSGNAADD1763 pKa = 4.19LSGMGFTDD1771 pKa = 4.03TLPAGLVIAATPAEE1785 pKa = 4.34TNTCGGSLTANAGASTFALSGGALAGGGTCAVSVTLRR1822 pKa = 11.84ALEE1825 pKa = 4.51AGSLTNPMFDD1835 pKa = 3.3LTSNIATATAASAILTVNAASAPVVAKK1862 pKa = 10.27GFAPAIINQGQTTTATVSIDD1882 pKa = 3.07NAANLIEE1889 pKa = 4.25ATGVAFTDD1897 pKa = 3.85ALPSGLVVAPTPAGSNTCGGTLTANAGASSFALSGGTIAEE1937 pKa = 4.42GASCQVTVALRR1948 pKa = 11.84ALDD1951 pKa = 4.43GGTLTNPAFDD1961 pKa = 3.88VTSSIATGTVAATDD1975 pKa = 3.76LTVNAAAAPGFAKK1988 pKa = 10.46AFSVSTLDD1996 pKa = 3.5QGQSLTATVTIDD2008 pKa = 3.32NAPNAIEE2015 pKa = 4.15ASNLAFTDD2023 pKa = 3.75ALPAGLVIGTASGGGSLISTTCGGTASATTGASSFSLSGGTLAEE2067 pKa = 4.56GASCTVVVVLRR2078 pKa = 11.84AVGAGTLTNPAMDD2091 pKa = 4.23LTSSIATATSAPVSLTANAAAAPVVTKK2118 pKa = 9.82VFAPATIEE2126 pKa = 3.89QGQTTTATITIDD2138 pKa = 3.15NAGNSIEE2145 pKa = 4.08AASLAFTDD2153 pKa = 4.09TLPAGLQVAAVPNEE2167 pKa = 3.97SNTCGGTLTAAAGTGAVSLSGGTLAEE2193 pKa = 4.49GASCAIAVDD2202 pKa = 4.69LRR2204 pKa = 11.84ATLPGTLTNPGFDD2217 pKa = 3.59VTSSIATGSAAATDD2231 pKa = 3.92LTVNAAAAMGFAKK2244 pKa = 10.4AFAPATIDD2252 pKa = 3.53LGSSSTLTFTIDD2264 pKa = 2.95NSANAIEE2271 pKa = 4.29AASMAFTDD2279 pKa = 4.28SFPAGLIVSSAPAATNTCGGTFTATALSGLVSLSGGTLAAGTTCTLSVQVQALTTGSLDD2338 pKa = 3.56NTSSEE2343 pKa = 4.36LTSDD2347 pKa = 4.37LPSASPVATASLTVNAVPITSAASFNPPIIEE2378 pKa = 3.98LFEE2381 pKa = 4.04LTEE2384 pKa = 4.78LVFLFQNDD2392 pKa = 3.17ADD2394 pKa = 3.98IDD2396 pKa = 3.94QILNFIQNLPAGLNVGGAATSRR2418 pKa = 11.84ANAMAGTNVSTTCSGGTVTAVEE2440 pKa = 5.06GSTQVQISGATVPANGSCEE2459 pKa = 4.13LKK2461 pKa = 11.15VNVIANAIGDD2471 pKa = 4.0FSNLVGDD2478 pKa = 4.33ADD2480 pKa = 4.2LVVNTIARR2488 pKa = 11.84AFVTYY2493 pKa = 10.6VVTGSDD2499 pKa = 2.95GSYY2502 pKa = 10.57AFSSSATEE2510 pKa = 3.9LNATVAVAGGTGTQGPLRR2528 pKa = 11.84SSEE2531 pKa = 3.65GSYY2534 pKa = 10.93SVAIAPPTGVAFDD2547 pKa = 4.9SGVCDD2552 pKa = 5.29DD2553 pKa = 5.04GDD2555 pKa = 4.22SSVDD2559 pKa = 3.63LANNAIALTLSPLEE2573 pKa = 4.34SVTCTFTALSPVQQTVDD2590 pKa = 3.88VINQFLTKK2598 pKa = 10.21RR2599 pKa = 11.84ADD2601 pKa = 4.26LILSTEE2607 pKa = 4.21PNLGARR2613 pKa = 11.84GRR2615 pKa = 11.84DD2616 pKa = 3.05RR2617 pKa = 11.84LGRR2620 pKa = 11.84GFGNASALSFANGDD2634 pKa = 4.04LKK2636 pKa = 11.71AMLPFTATMQPGNYY2650 pKa = 9.16SFKK2653 pKa = 11.03SSLLQVRR2660 pKa = 11.84QAAASVQLAHH2670 pKa = 7.21GSTKK2674 pKa = 10.67DD2675 pKa = 3.37AIYY2678 pKa = 10.31VPNYY2682 pKa = 10.41RR2683 pKa = 11.84FDD2685 pKa = 3.39AWFEE2689 pKa = 3.89AHH2691 pKa = 6.81HH2692 pKa = 6.64KK2693 pKa = 10.77EE2694 pKa = 4.86FDD2696 pKa = 3.18TGTNGTGHH2704 pKa = 6.04FTAAHH2709 pKa = 6.39IGADD2713 pKa = 3.43YY2714 pKa = 11.18VLNEE2718 pKa = 4.01NLLIGAMVSFDD2729 pKa = 4.05SMEE2732 pKa = 5.42DD2733 pKa = 3.24DD2734 pKa = 3.83TAISSASGSGWLIGPYY2750 pKa = 7.9MTAKK2754 pKa = 10.47LAPNLYY2760 pKa = 10.08FDD2762 pKa = 4.57ARR2764 pKa = 11.84LAGGASNNKK2773 pKa = 8.6VSPFNTYY2780 pKa = 9.01TDD2782 pKa = 3.41SFDD2785 pKa = 3.5TRR2787 pKa = 11.84RR2788 pKa = 11.84LLASASLSGDD2798 pKa = 3.5FQQGNWTISPTASLSYY2814 pKa = 10.56YY2815 pKa = 10.5RR2816 pKa = 11.84EE2817 pKa = 4.19TQDD2820 pKa = 4.61SYY2822 pKa = 11.69IDD2824 pKa = 3.55STGASIPSQTVEE2836 pKa = 4.41LGQLKK2841 pKa = 10.17IGPTFTGRR2849 pKa = 11.84FLGDD2853 pKa = 3.76DD2854 pKa = 4.05GEE2856 pKa = 5.58IIAPYY2861 pKa = 9.88FSVDD2865 pKa = 3.37AIYY2868 pKa = 11.21NLGQTSGVTVTNPDD2882 pKa = 3.47TASTSGWRR2890 pKa = 11.84GRR2892 pKa = 11.84LKK2894 pKa = 10.92AGVTVTEE2901 pKa = 4.23EE2902 pKa = 3.87NGTKK2906 pKa = 10.39LGIGATYY2913 pKa = 10.74DD2914 pKa = 3.27GLFRR2918 pKa = 11.84SDD2920 pKa = 3.01FDD2922 pKa = 3.17AWGLSFEE2929 pKa = 5.52LEE2931 pKa = 4.07IPLQKK2936 pKa = 9.32PTARR2940 pKa = 4.09
MM1 pKa = 7.82SIFSKK6 pKa = 10.83LNRR9 pKa = 11.84ALSAIFFVAVAAAGSPSWAQPAFTVGFTPNSIAPSGTTRR48 pKa = 11.84MIFTIDD54 pKa = 3.46NSSGGAVTSMDD65 pKa = 3.65FTNALPAGITVAVAPNAFTSCGGASASSATVTAMAGGSSIAMADD109 pKa = 3.62GQLAPGATCTVEE121 pKa = 4.45ANVTATATATNGPATLNTSNGSSASASVGLTVDD154 pKa = 3.8AAATAVGKK162 pKa = 10.54AFAPTTIGIGGVSRR176 pKa = 11.84LTLTVDD182 pKa = 3.55NSAGANAQNFTLTDD196 pKa = 3.64NLPAGMVLADD206 pKa = 4.47PPNTATTCPVAQSQTLTGVGGGSILEE232 pKa = 4.16LQSTGGLVFTPSFMVVNALPAGTVCTVSADD262 pKa = 3.55VTASVPGSLQNVTEE276 pKa = 4.34PFLDD280 pKa = 3.91SGSAAATLTVTGAPAGAPQLTKK302 pKa = 10.77EE303 pKa = 4.24FTDD306 pKa = 4.14DD307 pKa = 3.28PVAAGGSATLEE318 pKa = 3.78FEE320 pKa = 3.78ILNRR324 pKa = 11.84SRR326 pKa = 11.84SDD328 pKa = 3.08AASNIAFTDD337 pKa = 3.53DD338 pKa = 2.98MSALFGALAGTVTAIGTNTCGGTPSGVGTSSLSYY372 pKa = 10.75SGGTVGAGLRR382 pKa = 11.84CNITVTISVPAGVSTGSYY400 pKa = 9.24TNTSTAVTADD410 pKa = 3.48VGGMSLTGNAATANLSVLAALDD432 pKa = 4.11LPPVLSFTTPDD443 pKa = 3.23GNAGGVSTWSFTLSNAASSALTDD466 pKa = 3.42GTFDD470 pKa = 3.49VQMNPPLPFPTNASLPPMPDD490 pKa = 3.29PPCGVGSSLSLAFLDD505 pKa = 4.1TDD507 pKa = 3.68QQALRR512 pKa = 11.84LTGASLATSGAAGDD526 pKa = 3.88SCTFDD531 pKa = 3.27VDD533 pKa = 3.37IGLNSDD539 pKa = 5.14LPPLQYY545 pKa = 11.04NFTSGAPSGTVNGATRR561 pKa = 11.84TGTPASSSMTVGTNLNLSFAKK582 pKa = 10.45SFLATAVPGGTVDD595 pKa = 3.94LEE597 pKa = 4.28FVITSSGDD605 pKa = 3.46STDD608 pKa = 2.91ATAISFTDD616 pKa = 5.0DD617 pKa = 3.09INAMLAGTSLNAVISNEE634 pKa = 4.01CGGNLTGTTNLSYY647 pKa = 10.57TGGSLTGGTSCAIVTRR663 pKa = 11.84LDD665 pKa = 3.43VPAAAAIGTYY675 pKa = 10.32TNTTSALSATPSGSSTVEE693 pKa = 3.91TFGTASADD701 pKa = 3.47LTVAGLTFEE710 pKa = 5.78KK711 pKa = 10.68EE712 pKa = 4.04FLTNTVLAGDD722 pKa = 3.84TTTLRR727 pKa = 11.84FTISNVHH734 pKa = 5.72PTADD738 pKa = 3.23ATITSFTDD746 pKa = 3.15NLQQNLTGLAATGGPSGNTCGGTLSGTTFLIYY778 pKa = 10.26TGGSVTSGMSCTIDD792 pKa = 3.52VEE794 pKa = 4.53VLVPVGTADD803 pKa = 3.25GSYY806 pKa = 11.42GNTTSTLSATQGGGPVVVDD825 pKa = 4.0PATDD829 pKa = 3.14ILNVQGLQIEE839 pKa = 4.43LSKK842 pKa = 11.32AFTDD846 pKa = 4.42DD847 pKa = 3.45PVAPGDD853 pKa = 3.64NATLEE858 pKa = 4.41FTLSNLSSTEE868 pKa = 4.12AISNIAFTDD877 pKa = 3.5DD878 pKa = 3.76LAAMLPGTTFASVAGNDD895 pKa = 3.41CGATVGGTGTSTISVSGTSLAASATCTITVAVTVPGGAVDD935 pKa = 3.52NTYY938 pKa = 10.96TNTTSSVTGDD948 pKa = 2.79IGGLAVTGDD957 pKa = 3.84PATAQLSVSSILPLAFSKK975 pKa = 10.94SFSGSVASGEE985 pKa = 4.37SGTLTFTLSNPNTTALSDD1003 pKa = 4.2LGFTDD1008 pKa = 5.29DD1009 pKa = 4.78LNAMLTGVVATSLPSAPCGLGSSITGTSSLTASGFSLDD1047 pKa = 4.27ASGGSCTFDD1056 pKa = 2.78VGFSVPVTATGATYY1070 pKa = 11.21LNTTSEE1076 pKa = 4.37LVSFGLSLAPPATANLIVTDD1096 pKa = 4.52PVADD1100 pKa = 4.37LSLAIADD1107 pKa = 4.24SPDD1110 pKa = 3.59PVTPGQNLVYY1120 pKa = 10.12TVTASNAGPQTAQSVAVNLPVPTGTTLVSTTGCAEE1155 pKa = 4.41DD1156 pKa = 3.86PSGAATCSLGNIASGGNAQYY1176 pKa = 10.53TLTVTVDD1183 pKa = 3.0AATSGTVSASGTVSSSTSDD1202 pKa = 3.19PVAGNNTATASTSAPATADD1221 pKa = 3.12ISITKK1226 pKa = 10.16SDD1228 pKa = 3.63GVTSANSGGTTTYY1241 pKa = 10.78SIVVANAGPSADD1253 pKa = 3.74PAVSVADD1260 pKa = 3.92TFAADD1265 pKa = 4.66LGCATTSVAAGGATGNTAAFSGDD1288 pKa = 3.07IAEE1291 pKa = 4.46TLSMPASSSVTYY1303 pKa = 9.09TATCDD1308 pKa = 3.17IAITATGTLSNTATATASVLDD1329 pKa = 4.37PNATNNSATDD1339 pKa = 3.17SDD1341 pKa = 4.34TALVAPVAMGFSKK1354 pKa = 10.76AFSPATIDD1362 pKa = 3.19QGANTTLTFTIDD1374 pKa = 3.04NTANAVAADD1383 pKa = 3.92NLNFTDD1389 pKa = 5.15GFPLGVDD1396 pKa = 3.92LSVAAPVVSSSTCGGTFNPVAGGGNLVFSGGSVAAGATCTVSVLLNARR1444 pKa = 11.84GSGTATNTSSQLGSFFPTPSAAATADD1470 pKa = 3.23ITVNPAVAPTFAKK1483 pKa = 10.65AFGAATIEE1491 pKa = 4.06QGQTTTATFTINNSANSIEE1510 pKa = 4.14ASSLQFSDD1518 pKa = 3.71TLPVEE1523 pKa = 4.44LVVAATPNASTTCTGGTITATAGSGSILYY1552 pKa = 10.56SGGEE1556 pKa = 4.03VAAGATCTVTVTVDD1570 pKa = 3.32SLGSGSIANPSLNLTSSLPTATAAATPLTINPATAPGVAKK1610 pKa = 9.81TFTPSAITQGEE1621 pKa = 4.31TSALSITIDD1630 pKa = 3.12NSANFLEE1637 pKa = 4.45MTGVAFTDD1645 pKa = 4.08AFPSGMVVGPTPGASNTCGGTFSASAGSGTISLSGGTVAAGATCSVGVTVRR1696 pKa = 11.84GNAAGTLTNPAFDD1709 pKa = 3.67VTSNIATGTAAATDD1723 pKa = 4.08LAVNPAAAPGFAKK1736 pKa = 10.65AFGAGTINQGGTVVATVSVDD1756 pKa = 3.39NSGNAADD1763 pKa = 4.19LSGMGFTDD1771 pKa = 4.03TLPAGLVIAATPAEE1785 pKa = 4.34TNTCGGSLTANAGASTFALSGGALAGGGTCAVSVTLRR1822 pKa = 11.84ALEE1825 pKa = 4.51AGSLTNPMFDD1835 pKa = 3.3LTSNIATATAASAILTVNAASAPVVAKK1862 pKa = 10.27GFAPAIINQGQTTTATVSIDD1882 pKa = 3.07NAANLIEE1889 pKa = 4.25ATGVAFTDD1897 pKa = 3.85ALPSGLVVAPTPAGSNTCGGTLTANAGASSFALSGGTIAEE1937 pKa = 4.42GASCQVTVALRR1948 pKa = 11.84ALDD1951 pKa = 4.43GGTLTNPAFDD1961 pKa = 3.88VTSSIATGTVAATDD1975 pKa = 3.76LTVNAAAAPGFAKK1988 pKa = 10.46AFSVSTLDD1996 pKa = 3.5QGQSLTATVTIDD2008 pKa = 3.32NAPNAIEE2015 pKa = 4.15ASNLAFTDD2023 pKa = 3.75ALPAGLVIGTASGGGSLISTTCGGTASATTGASSFSLSGGTLAEE2067 pKa = 4.56GASCTVVVVLRR2078 pKa = 11.84AVGAGTLTNPAMDD2091 pKa = 4.23LTSSIATATSAPVSLTANAAAAPVVTKK2118 pKa = 9.82VFAPATIEE2126 pKa = 3.89QGQTTTATITIDD2138 pKa = 3.15NAGNSIEE2145 pKa = 4.08AASLAFTDD2153 pKa = 4.09TLPAGLQVAAVPNEE2167 pKa = 3.97SNTCGGTLTAAAGTGAVSLSGGTLAEE2193 pKa = 4.49GASCAIAVDD2202 pKa = 4.69LRR2204 pKa = 11.84ATLPGTLTNPGFDD2217 pKa = 3.59VTSSIATGSAAATDD2231 pKa = 3.92LTVNAAAAMGFAKK2244 pKa = 10.4AFAPATIDD2252 pKa = 3.53LGSSSTLTFTIDD2264 pKa = 2.95NSANAIEE2271 pKa = 4.29AASMAFTDD2279 pKa = 4.28SFPAGLIVSSAPAATNTCGGTFTATALSGLVSLSGGTLAAGTTCTLSVQVQALTTGSLDD2338 pKa = 3.56NTSSEE2343 pKa = 4.36LTSDD2347 pKa = 4.37LPSASPVATASLTVNAVPITSAASFNPPIIEE2378 pKa = 3.98LFEE2381 pKa = 4.04LTEE2384 pKa = 4.78LVFLFQNDD2392 pKa = 3.17ADD2394 pKa = 3.98IDD2396 pKa = 3.94QILNFIQNLPAGLNVGGAATSRR2418 pKa = 11.84ANAMAGTNVSTTCSGGTVTAVEE2440 pKa = 5.06GSTQVQISGATVPANGSCEE2459 pKa = 4.13LKK2461 pKa = 11.15VNVIANAIGDD2471 pKa = 4.0FSNLVGDD2478 pKa = 4.33ADD2480 pKa = 4.2LVVNTIARR2488 pKa = 11.84AFVTYY2493 pKa = 10.6VVTGSDD2499 pKa = 2.95GSYY2502 pKa = 10.57AFSSSATEE2510 pKa = 3.9LNATVAVAGGTGTQGPLRR2528 pKa = 11.84SSEE2531 pKa = 3.65GSYY2534 pKa = 10.93SVAIAPPTGVAFDD2547 pKa = 4.9SGVCDD2552 pKa = 5.29DD2553 pKa = 5.04GDD2555 pKa = 4.22SSVDD2559 pKa = 3.63LANNAIALTLSPLEE2573 pKa = 4.34SVTCTFTALSPVQQTVDD2590 pKa = 3.88VINQFLTKK2598 pKa = 10.21RR2599 pKa = 11.84ADD2601 pKa = 4.26LILSTEE2607 pKa = 4.21PNLGARR2613 pKa = 11.84GRR2615 pKa = 11.84DD2616 pKa = 3.05RR2617 pKa = 11.84LGRR2620 pKa = 11.84GFGNASALSFANGDD2634 pKa = 4.04LKK2636 pKa = 11.71AMLPFTATMQPGNYY2650 pKa = 9.16SFKK2653 pKa = 11.03SSLLQVRR2660 pKa = 11.84QAAASVQLAHH2670 pKa = 7.21GSTKK2674 pKa = 10.67DD2675 pKa = 3.37AIYY2678 pKa = 10.31VPNYY2682 pKa = 10.41RR2683 pKa = 11.84FDD2685 pKa = 3.39AWFEE2689 pKa = 3.89AHH2691 pKa = 6.81HH2692 pKa = 6.64KK2693 pKa = 10.77EE2694 pKa = 4.86FDD2696 pKa = 3.18TGTNGTGHH2704 pKa = 6.04FTAAHH2709 pKa = 6.39IGADD2713 pKa = 3.43YY2714 pKa = 11.18VLNEE2718 pKa = 4.01NLLIGAMVSFDD2729 pKa = 4.05SMEE2732 pKa = 5.42DD2733 pKa = 3.24DD2734 pKa = 3.83TAISSASGSGWLIGPYY2750 pKa = 7.9MTAKK2754 pKa = 10.47LAPNLYY2760 pKa = 10.08FDD2762 pKa = 4.57ARR2764 pKa = 11.84LAGGASNNKK2773 pKa = 8.6VSPFNTYY2780 pKa = 9.01TDD2782 pKa = 3.41SFDD2785 pKa = 3.5TRR2787 pKa = 11.84RR2788 pKa = 11.84LLASASLSGDD2798 pKa = 3.5FQQGNWTISPTASLSYY2814 pKa = 10.56YY2815 pKa = 10.5RR2816 pKa = 11.84EE2817 pKa = 4.19TQDD2820 pKa = 4.61SYY2822 pKa = 11.69IDD2824 pKa = 3.55STGASIPSQTVEE2836 pKa = 4.41LGQLKK2841 pKa = 10.17IGPTFTGRR2849 pKa = 11.84FLGDD2853 pKa = 3.76DD2854 pKa = 4.05GEE2856 pKa = 5.58IIAPYY2861 pKa = 9.88FSVDD2865 pKa = 3.37AIYY2868 pKa = 11.21NLGQTSGVTVTNPDD2882 pKa = 3.47TASTSGWRR2890 pKa = 11.84GRR2892 pKa = 11.84LKK2894 pKa = 10.92AGVTVTEE2901 pKa = 4.23EE2902 pKa = 3.87NGTKK2906 pKa = 10.39LGIGATYY2913 pKa = 10.74DD2914 pKa = 3.27GLFRR2918 pKa = 11.84SDD2920 pKa = 3.01FDD2922 pKa = 3.17AWGLSFEE2929 pKa = 5.52LEE2931 pKa = 4.07IPLQKK2936 pKa = 9.32PTARR2940 pKa = 4.09
Molecular weight: 287.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0J9H4A4|A0A0J9H4A4_9RHOB Membrane protein OS=Candidatus Rhodobacter lobularis OX=1675527 GN=AIOL_000659 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1428562 |
37 |
2940 |
299.9 |
32.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.623 ± 0.051 | 0.942 ± 0.01 |
5.927 ± 0.031 | 5.974 ± 0.034 |
3.832 ± 0.022 | 8.847 ± 0.039 |
2.074 ± 0.018 | 5.071 ± 0.027 |
3.098 ± 0.029 | 9.885 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.777 ± 0.017 | 2.673 ± 0.021 |
5.053 ± 0.026 | 3.367 ± 0.021 |
6.51 ± 0.037 | 5.076 ± 0.03 |
5.25 ± 0.029 | 7.257 ± 0.032 |
1.481 ± 0.014 | 2.283 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
