
Paramuricea placomus associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 9.13
Get precalculated fractions of proteins
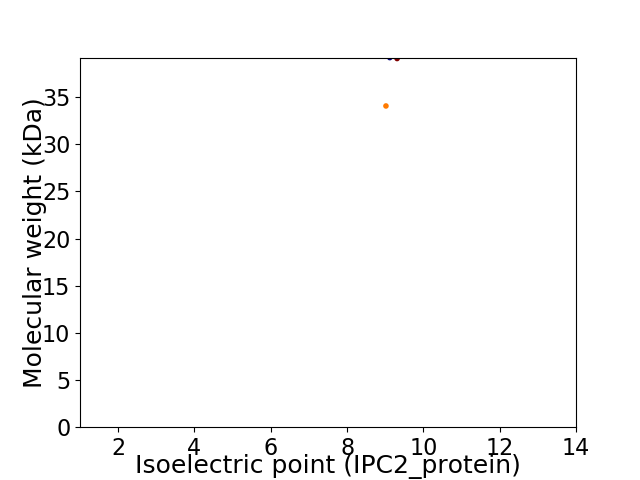
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLR9|A0A0K1RLR9_9CIRC Putative capsid protein OS=Paramuricea placomus associated circular virus OX=1692260 PE=4 SV=1
MM1 pKa = 7.31FGEE4 pKa = 4.16ADD6 pKa = 3.15EE7 pKa = 4.89GEE9 pKa = 4.4RR10 pKa = 11.84ANLDD14 pKa = 3.03IEE16 pKa = 4.39PPKK19 pKa = 10.28KK20 pKa = 9.84RR21 pKa = 11.84KK22 pKa = 9.76KK23 pKa = 9.19RR24 pKa = 11.84DD25 pKa = 2.96HH26 pKa = 5.99RR27 pKa = 11.84AEE29 pKa = 4.1GKK31 pKa = 9.96RR32 pKa = 11.84RR33 pKa = 11.84ALRR36 pKa = 11.84YY37 pKa = 9.39RR38 pKa = 11.84NWVFTLNNPQEE49 pKa = 4.39HH50 pKa = 6.51GFPRR54 pKa = 11.84LTSDD58 pKa = 4.62MIWLRR63 pKa = 11.84YY64 pKa = 9.03GEE66 pKa = 4.54EE67 pKa = 4.09VAPTTGTPHH76 pKa = 6.02LQGVVHH82 pKa = 6.68FKK84 pKa = 10.51SKK86 pKa = 10.47RR87 pKa = 11.84VMPTDD92 pKa = 4.16LYY94 pKa = 8.67PWCRR98 pKa = 11.84NAHH101 pKa = 5.34WEE103 pKa = 4.2HH104 pKa = 6.21MGGTISANLAYY115 pKa = 9.34TGKK118 pKa = 9.9EE119 pKa = 3.91ANEE122 pKa = 4.09EE123 pKa = 3.95AGTLHH128 pKa = 6.38EE129 pKa = 4.86FGVRR133 pKa = 11.84PLSNKK138 pKa = 7.19EE139 pKa = 3.78ARR141 pKa = 11.84EE142 pKa = 3.88KK143 pKa = 10.81GAAATKK149 pKa = 9.77EE150 pKa = 3.92RR151 pKa = 11.84YY152 pKa = 9.6INILAHH158 pKa = 6.51AKK160 pKa = 10.35AGEE163 pKa = 3.82FDD165 pKa = 3.78KK166 pKa = 11.68LEE168 pKa = 4.06EE169 pKa = 4.44EE170 pKa = 4.69YY171 pKa = 10.54PGDD174 pKa = 3.91FLRR177 pKa = 11.84MYY179 pKa = 8.77KK180 pKa = 9.15TLHH183 pKa = 6.83CIRR186 pKa = 11.84DD187 pKa = 3.73GAQGRR192 pKa = 11.84SEE194 pKa = 4.33PIARR198 pKa = 11.84LEE200 pKa = 4.06HH201 pKa = 5.45WWIVGPTGCGKK212 pKa = 10.24SRR214 pKa = 11.84AVWEE218 pKa = 4.4TVGASQLYY226 pKa = 10.03SKK228 pKa = 10.75LLRR231 pKa = 11.84TSGGTTTTTKK241 pKa = 9.62TGSYY245 pKa = 8.55STTILPYY252 pKa = 10.75GKK254 pKa = 8.09TKK256 pKa = 10.12PVSKK260 pKa = 9.93IGRR263 pKa = 11.84TTIPSLRR270 pKa = 11.84RR271 pKa = 11.84SRR273 pKa = 11.84ALQKK277 pKa = 10.76EE278 pKa = 4.3SAQLISSSLATTASQTDD295 pKa = 3.4SSSPMMSSQQ304 pKa = 3.35
MM1 pKa = 7.31FGEE4 pKa = 4.16ADD6 pKa = 3.15EE7 pKa = 4.89GEE9 pKa = 4.4RR10 pKa = 11.84ANLDD14 pKa = 3.03IEE16 pKa = 4.39PPKK19 pKa = 10.28KK20 pKa = 9.84RR21 pKa = 11.84KK22 pKa = 9.76KK23 pKa = 9.19RR24 pKa = 11.84DD25 pKa = 2.96HH26 pKa = 5.99RR27 pKa = 11.84AEE29 pKa = 4.1GKK31 pKa = 9.96RR32 pKa = 11.84RR33 pKa = 11.84ALRR36 pKa = 11.84YY37 pKa = 9.39RR38 pKa = 11.84NWVFTLNNPQEE49 pKa = 4.39HH50 pKa = 6.51GFPRR54 pKa = 11.84LTSDD58 pKa = 4.62MIWLRR63 pKa = 11.84YY64 pKa = 9.03GEE66 pKa = 4.54EE67 pKa = 4.09VAPTTGTPHH76 pKa = 6.02LQGVVHH82 pKa = 6.68FKK84 pKa = 10.51SKK86 pKa = 10.47RR87 pKa = 11.84VMPTDD92 pKa = 4.16LYY94 pKa = 8.67PWCRR98 pKa = 11.84NAHH101 pKa = 5.34WEE103 pKa = 4.2HH104 pKa = 6.21MGGTISANLAYY115 pKa = 9.34TGKK118 pKa = 9.9EE119 pKa = 3.91ANEE122 pKa = 4.09EE123 pKa = 3.95AGTLHH128 pKa = 6.38EE129 pKa = 4.86FGVRR133 pKa = 11.84PLSNKK138 pKa = 7.19EE139 pKa = 3.78ARR141 pKa = 11.84EE142 pKa = 3.88KK143 pKa = 10.81GAAATKK149 pKa = 9.77EE150 pKa = 3.92RR151 pKa = 11.84YY152 pKa = 9.6INILAHH158 pKa = 6.51AKK160 pKa = 10.35AGEE163 pKa = 3.82FDD165 pKa = 3.78KK166 pKa = 11.68LEE168 pKa = 4.06EE169 pKa = 4.44EE170 pKa = 4.69YY171 pKa = 10.54PGDD174 pKa = 3.91FLRR177 pKa = 11.84MYY179 pKa = 8.77KK180 pKa = 9.15TLHH183 pKa = 6.83CIRR186 pKa = 11.84DD187 pKa = 3.73GAQGRR192 pKa = 11.84SEE194 pKa = 4.33PIARR198 pKa = 11.84LEE200 pKa = 4.06HH201 pKa = 5.45WWIVGPTGCGKK212 pKa = 10.24SRR214 pKa = 11.84AVWEE218 pKa = 4.4TVGASQLYY226 pKa = 10.03SKK228 pKa = 10.75LLRR231 pKa = 11.84TSGGTTTTTKK241 pKa = 9.62TGSYY245 pKa = 8.55STTILPYY252 pKa = 10.75GKK254 pKa = 8.09TKK256 pKa = 10.12PVSKK260 pKa = 9.93IGRR263 pKa = 11.84TTIPSLRR270 pKa = 11.84RR271 pKa = 11.84SRR273 pKa = 11.84ALQKK277 pKa = 10.76EE278 pKa = 4.3SAQLISSSLATTASQTDD295 pKa = 3.4SSSPMMSSQQ304 pKa = 3.35
Molecular weight: 34.05 kDa
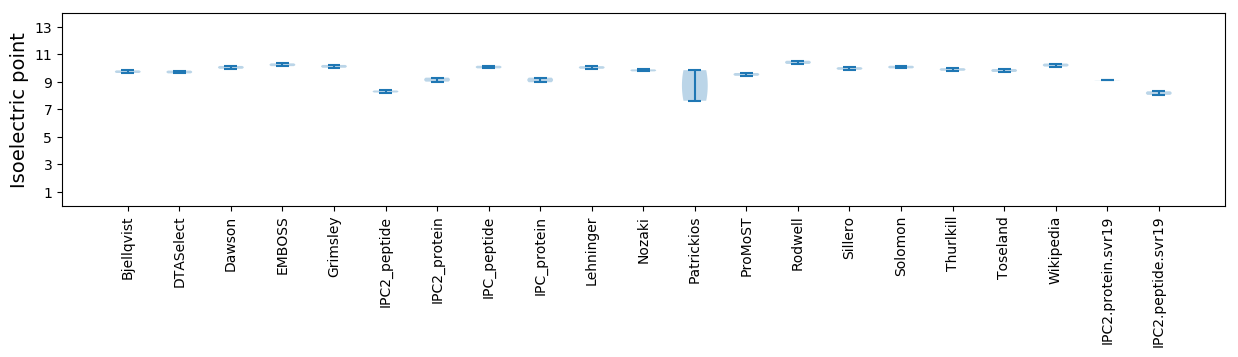
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLR9|A0A0K1RLR9_9CIRC Putative capsid protein OS=Paramuricea placomus associated circular virus OX=1692260 PE=4 SV=1
MM1 pKa = 7.63AYY3 pKa = 9.58VRR5 pKa = 11.84KK6 pKa = 9.92RR7 pKa = 11.84SFRR10 pKa = 11.84KK11 pKa = 7.57RR12 pKa = 11.84TSWKK16 pKa = 9.94KK17 pKa = 4.82MTRR20 pKa = 11.84GFRR23 pKa = 11.84KK24 pKa = 9.75KK25 pKa = 10.4KK26 pKa = 8.46PAKK29 pKa = 10.51ANLNWRR35 pKa = 11.84KK36 pKa = 9.41RR37 pKa = 11.84RR38 pKa = 11.84SQSFRR43 pKa = 11.84RR44 pKa = 11.84ARR46 pKa = 11.84RR47 pKa = 11.84TRR49 pKa = 11.84RR50 pKa = 11.84AANPGTAIVTSSTFEE65 pKa = 3.85MNFAGTSPILGILNGTNLAEE85 pKa = 4.21MGGVASCLVRR95 pKa = 11.84EE96 pKa = 4.99LYY98 pKa = 9.99IQPGRR103 pKa = 11.84IAGLLARR110 pKa = 11.84QFVYY114 pKa = 11.06NMYY117 pKa = 10.23KK118 pKa = 9.91IYY120 pKa = 10.48SVEE123 pKa = 4.02YY124 pKa = 10.39KK125 pKa = 10.65LIRR128 pKa = 11.84SDD130 pKa = 3.18KK131 pKa = 10.51SRR133 pKa = 11.84ISSGPISADD142 pKa = 3.07YY143 pKa = 10.63AYY145 pKa = 10.72ADD147 pKa = 3.86FAMLMPNTHH156 pKa = 6.35NEE158 pKa = 4.04DD159 pKa = 3.54FPGISTAGSIQTTRR173 pKa = 11.84KK174 pKa = 9.53LMNWMLQQKK183 pKa = 8.92GSKK186 pKa = 7.93MVKK189 pKa = 9.38LQQNFAKK196 pKa = 9.51MKK198 pKa = 8.91VPAMVNVEE206 pKa = 5.27RR207 pKa = 11.84EE208 pKa = 4.14FQQNTGTGTVTQMSTIPHH226 pKa = 6.37PWIEE230 pKa = 3.95IDD232 pKa = 3.43TDD234 pKa = 3.48KK235 pKa = 11.04MNTMNIGQVIFAMPALNISTFYY257 pKa = 10.66PVFPTPEE264 pKa = 3.97AQTAAGTGDD273 pKa = 3.34VGAPGQTYY281 pKa = 11.19DD282 pKa = 4.3NLKK285 pKa = 10.73KK286 pKa = 10.21MYY288 pKa = 9.56DD289 pKa = 3.49YY290 pKa = 11.26SIFATVRR297 pKa = 11.84WGVKK301 pKa = 9.4GKK303 pKa = 10.94YY304 pKa = 9.11IDD306 pKa = 3.72PAVAQEE312 pKa = 3.84YY313 pKa = 8.8HH314 pKa = 6.19QPSQVGDD321 pKa = 3.98DD322 pKa = 3.74EE323 pKa = 4.89PEE325 pKa = 3.95EE326 pKa = 4.24FGEE329 pKa = 4.48DD330 pKa = 3.6TVDD333 pKa = 3.19MGSLIDD339 pKa = 4.06NMQNMTIAA347 pKa = 3.88
MM1 pKa = 7.63AYY3 pKa = 9.58VRR5 pKa = 11.84KK6 pKa = 9.92RR7 pKa = 11.84SFRR10 pKa = 11.84KK11 pKa = 7.57RR12 pKa = 11.84TSWKK16 pKa = 9.94KK17 pKa = 4.82MTRR20 pKa = 11.84GFRR23 pKa = 11.84KK24 pKa = 9.75KK25 pKa = 10.4KK26 pKa = 8.46PAKK29 pKa = 10.51ANLNWRR35 pKa = 11.84KK36 pKa = 9.41RR37 pKa = 11.84RR38 pKa = 11.84SQSFRR43 pKa = 11.84RR44 pKa = 11.84ARR46 pKa = 11.84RR47 pKa = 11.84TRR49 pKa = 11.84RR50 pKa = 11.84AANPGTAIVTSSTFEE65 pKa = 3.85MNFAGTSPILGILNGTNLAEE85 pKa = 4.21MGGVASCLVRR95 pKa = 11.84EE96 pKa = 4.99LYY98 pKa = 9.99IQPGRR103 pKa = 11.84IAGLLARR110 pKa = 11.84QFVYY114 pKa = 11.06NMYY117 pKa = 10.23KK118 pKa = 9.91IYY120 pKa = 10.48SVEE123 pKa = 4.02YY124 pKa = 10.39KK125 pKa = 10.65LIRR128 pKa = 11.84SDD130 pKa = 3.18KK131 pKa = 10.51SRR133 pKa = 11.84ISSGPISADD142 pKa = 3.07YY143 pKa = 10.63AYY145 pKa = 10.72ADD147 pKa = 3.86FAMLMPNTHH156 pKa = 6.35NEE158 pKa = 4.04DD159 pKa = 3.54FPGISTAGSIQTTRR173 pKa = 11.84KK174 pKa = 9.53LMNWMLQQKK183 pKa = 8.92GSKK186 pKa = 7.93MVKK189 pKa = 9.38LQQNFAKK196 pKa = 9.51MKK198 pKa = 8.91VPAMVNVEE206 pKa = 5.27RR207 pKa = 11.84EE208 pKa = 4.14FQQNTGTGTVTQMSTIPHH226 pKa = 6.37PWIEE230 pKa = 3.95IDD232 pKa = 3.43TDD234 pKa = 3.48KK235 pKa = 11.04MNTMNIGQVIFAMPALNISTFYY257 pKa = 10.66PVFPTPEE264 pKa = 3.97AQTAAGTGDD273 pKa = 3.34VGAPGQTYY281 pKa = 11.19DD282 pKa = 4.3NLKK285 pKa = 10.73KK286 pKa = 10.21MYY288 pKa = 9.56DD289 pKa = 3.49YY290 pKa = 11.26SIFATVRR297 pKa = 11.84WGVKK301 pKa = 9.4GKK303 pKa = 10.94YY304 pKa = 9.11IDD306 pKa = 3.72PAVAQEE312 pKa = 3.84YY313 pKa = 8.8HH314 pKa = 6.19QPSQVGDD321 pKa = 3.98DD322 pKa = 3.74EE323 pKa = 4.89PEE325 pKa = 3.95EE326 pKa = 4.24FGEE329 pKa = 4.48DD330 pKa = 3.6TVDD333 pKa = 3.19MGSLIDD339 pKa = 4.06NMQNMTIAA347 pKa = 3.88
Molecular weight: 39.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
651 |
304 |
347 |
325.5 |
36.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.295 ± 0.049 | 0.614 ± 0.258 |
3.687 ± 0.504 | 5.837 ± 1.427 |
3.379 ± 0.747 | 7.834 ± 0.498 |
1.997 ± 0.896 | 5.069 ± 0.778 |
6.759 ± 0.103 | 5.991 ± 1.092 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.147 ± 1.279 | 4.301 ± 0.93 |
5.223 ± 0.028 | 3.994 ± 0.945 |
7.373 ± 0.59 | 7.066 ± 0.575 |
8.449 ± 0.528 | 4.455 ± 0.808 |
1.843 ± 0.319 | 3.687 ± 0.275 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
