
Hubei virga-like virus 21
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
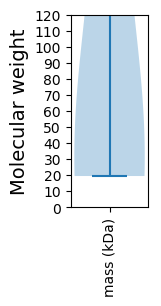
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KJW8|A0A1L3KJW8_9VIRU Uncharacterized protein OS=Hubei virga-like virus 21 OX=1923336 PE=4 SV=1
MM1 pKa = 7.62SLEE4 pKa = 4.13WCCILALLSCRR15 pKa = 11.84LRR17 pKa = 11.84QLVLVSATPLVHH29 pKa = 7.44RR30 pKa = 11.84RR31 pKa = 11.84IQPLVTFSGVEE42 pKa = 3.74PEE44 pKa = 4.01YY45 pKa = 10.84TLLVDD50 pKa = 3.9HH51 pKa = 6.83LPVVTEE57 pKa = 4.13GEE59 pKa = 4.17KK60 pKa = 11.01CLVAVAGDD68 pKa = 3.82SCEE71 pKa = 4.35LPDD74 pKa = 3.47YY75 pKa = 10.26CGQVEE80 pKa = 4.37RR81 pKa = 11.84FHH83 pKa = 6.89MKK85 pKa = 10.03VQDD88 pKa = 4.21LLLVICAGHH97 pKa = 6.35HH98 pKa = 5.42VLPGHH103 pKa = 6.6KK104 pKa = 9.66SDD106 pKa = 3.86IEE108 pKa = 3.84IFRR111 pKa = 11.84AYY113 pKa = 10.76SLDD116 pKa = 3.18ARR118 pKa = 11.84GEE120 pKa = 4.22FIPVEE125 pKa = 4.17LYY127 pKa = 10.91FIDD130 pKa = 5.02VYY132 pKa = 11.58SLDD135 pKa = 3.12RR136 pKa = 11.84KK137 pKa = 9.54YY138 pKa = 10.61RR139 pKa = 11.84YY140 pKa = 9.89SVVLPWAMLSKK151 pKa = 10.64LVVKK155 pKa = 8.72TAVQHH160 pKa = 5.66SFYY163 pKa = 10.78VDD165 pKa = 2.97KK166 pKa = 10.89DD167 pKa = 3.37LRR169 pKa = 11.84ICRR172 pKa = 11.84RR173 pKa = 11.84GTYY176 pKa = 9.68LRR178 pKa = 11.84SLTRR182 pKa = 11.84LLSRR186 pKa = 11.84RR187 pKa = 11.84ILNSEE192 pKa = 3.77KK193 pKa = 10.23QLYY196 pKa = 9.45PGDD199 pKa = 4.08APFLEE204 pKa = 4.68ANSLRR209 pKa = 11.84GASCRR214 pKa = 11.84PSGTHH219 pKa = 5.89EE220 pKa = 4.75PYY222 pKa = 10.58SCTDD226 pKa = 4.54LIVVNEE232 pKa = 4.47TYY234 pKa = 10.98ALCSSVWTQLHH245 pKa = 6.7PGACPMDD252 pKa = 3.44RR253 pKa = 11.84QYY255 pKa = 11.66RR256 pKa = 11.84RR257 pKa = 11.84GEE259 pKa = 3.84FGFEE263 pKa = 3.89DD264 pKa = 4.09VCTNITVLEE273 pKa = 4.13KK274 pKa = 10.74KK275 pKa = 10.08KK276 pKa = 11.0AFGEE280 pKa = 4.38DD281 pKa = 4.12PDD283 pKa = 5.68DD284 pKa = 3.8GWLQRR289 pKa = 11.84SLKK292 pKa = 10.35QVVNWLTTKK301 pKa = 10.12IDD303 pKa = 3.74DD304 pKa = 3.56FAEE307 pKa = 5.04FIEE310 pKa = 4.7GLFLKK315 pKa = 10.74LLEE318 pKa = 4.15KK319 pKa = 10.82VIAFMFSQLEE329 pKa = 4.21VLDD332 pKa = 4.17SLVEE336 pKa = 4.45FVDD339 pKa = 3.11SRR341 pKa = 11.84YY342 pKa = 9.99VVFEE346 pKa = 4.08LMVVSFIICYY356 pKa = 10.1RR357 pKa = 11.84SNLPAALIFVVIFGVTCGYY376 pKa = 11.06DD377 pKa = 3.0RR378 pKa = 11.84TRR380 pKa = 11.84DD381 pKa = 3.69FRR383 pKa = 11.84LLPEE387 pKa = 4.84LKK389 pKa = 10.52ALLLWSGG396 pKa = 3.46
MM1 pKa = 7.62SLEE4 pKa = 4.13WCCILALLSCRR15 pKa = 11.84LRR17 pKa = 11.84QLVLVSATPLVHH29 pKa = 7.44RR30 pKa = 11.84RR31 pKa = 11.84IQPLVTFSGVEE42 pKa = 3.74PEE44 pKa = 4.01YY45 pKa = 10.84TLLVDD50 pKa = 3.9HH51 pKa = 6.83LPVVTEE57 pKa = 4.13GEE59 pKa = 4.17KK60 pKa = 11.01CLVAVAGDD68 pKa = 3.82SCEE71 pKa = 4.35LPDD74 pKa = 3.47YY75 pKa = 10.26CGQVEE80 pKa = 4.37RR81 pKa = 11.84FHH83 pKa = 6.89MKK85 pKa = 10.03VQDD88 pKa = 4.21LLLVICAGHH97 pKa = 6.35HH98 pKa = 5.42VLPGHH103 pKa = 6.6KK104 pKa = 9.66SDD106 pKa = 3.86IEE108 pKa = 3.84IFRR111 pKa = 11.84AYY113 pKa = 10.76SLDD116 pKa = 3.18ARR118 pKa = 11.84GEE120 pKa = 4.22FIPVEE125 pKa = 4.17LYY127 pKa = 10.91FIDD130 pKa = 5.02VYY132 pKa = 11.58SLDD135 pKa = 3.12RR136 pKa = 11.84KK137 pKa = 9.54YY138 pKa = 10.61RR139 pKa = 11.84YY140 pKa = 9.89SVVLPWAMLSKK151 pKa = 10.64LVVKK155 pKa = 8.72TAVQHH160 pKa = 5.66SFYY163 pKa = 10.78VDD165 pKa = 2.97KK166 pKa = 10.89DD167 pKa = 3.37LRR169 pKa = 11.84ICRR172 pKa = 11.84RR173 pKa = 11.84GTYY176 pKa = 9.68LRR178 pKa = 11.84SLTRR182 pKa = 11.84LLSRR186 pKa = 11.84RR187 pKa = 11.84ILNSEE192 pKa = 3.77KK193 pKa = 10.23QLYY196 pKa = 9.45PGDD199 pKa = 4.08APFLEE204 pKa = 4.68ANSLRR209 pKa = 11.84GASCRR214 pKa = 11.84PSGTHH219 pKa = 5.89EE220 pKa = 4.75PYY222 pKa = 10.58SCTDD226 pKa = 4.54LIVVNEE232 pKa = 4.47TYY234 pKa = 10.98ALCSSVWTQLHH245 pKa = 6.7PGACPMDD252 pKa = 3.44RR253 pKa = 11.84QYY255 pKa = 11.66RR256 pKa = 11.84RR257 pKa = 11.84GEE259 pKa = 3.84FGFEE263 pKa = 3.89DD264 pKa = 4.09VCTNITVLEE273 pKa = 4.13KK274 pKa = 10.74KK275 pKa = 10.08KK276 pKa = 11.0AFGEE280 pKa = 4.38DD281 pKa = 4.12PDD283 pKa = 5.68DD284 pKa = 3.8GWLQRR289 pKa = 11.84SLKK292 pKa = 10.35QVVNWLTTKK301 pKa = 10.12IDD303 pKa = 3.74DD304 pKa = 3.56FAEE307 pKa = 5.04FIEE310 pKa = 4.7GLFLKK315 pKa = 10.74LLEE318 pKa = 4.15KK319 pKa = 10.82VIAFMFSQLEE329 pKa = 4.21VLDD332 pKa = 4.17SLVEE336 pKa = 4.45FVDD339 pKa = 3.11SRR341 pKa = 11.84YY342 pKa = 9.99VVFEE346 pKa = 4.08LMVVSFIICYY356 pKa = 10.1RR357 pKa = 11.84SNLPAALIFVVIFGVTCGYY376 pKa = 11.06DD377 pKa = 3.0RR378 pKa = 11.84TRR380 pKa = 11.84DD381 pKa = 3.69FRR383 pKa = 11.84LLPEE387 pKa = 4.84LKK389 pKa = 10.52ALLLWSGG396 pKa = 3.46
Molecular weight: 45.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJV3|A0A1L3KJV3_9VIRU Uncharacterized protein OS=Hubei virga-like virus 21 OX=1923336 PE=4 SV=1
MM1 pKa = 7.22VNGAFRR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84ARR12 pKa = 11.84ATVARR17 pKa = 11.84RR18 pKa = 11.84SFLDD22 pKa = 3.1SLLVVLLRR30 pKa = 11.84VVAHH34 pKa = 6.11PVSLVLAVFLVLFVAAEE51 pKa = 4.18VLEE54 pKa = 4.46TTGPLEE60 pKa = 4.97SLDD63 pKa = 3.96KK64 pKa = 11.03LIKK67 pKa = 10.19QEE69 pKa = 5.06LGSKK73 pKa = 10.14DD74 pKa = 3.33INSLEE79 pKa = 3.95KK80 pKa = 10.5FLLKK84 pKa = 10.91GFDD87 pKa = 3.16KK88 pKa = 11.24CIVFVILYY96 pKa = 5.85KK97 pKa = 9.98TKK99 pKa = 10.5VVATLAYY106 pKa = 10.31SIFVALNPTKK116 pKa = 10.59LRR118 pKa = 11.84WSVFGAAVLIVVAIPSLPVFYY139 pKa = 10.59HH140 pKa = 7.08IITAVALVFYY150 pKa = 10.53LALQRR155 pKa = 11.84IEE157 pKa = 4.99HH158 pKa = 6.57KK159 pKa = 10.63ALTVFIYY166 pKa = 9.97VAGMVLYY173 pKa = 9.03TSAVVVSVATAGAGKK188 pKa = 10.14RR189 pKa = 11.84NSTGAPP195 pKa = 3.29
MM1 pKa = 7.22VNGAFRR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84ARR12 pKa = 11.84ATVARR17 pKa = 11.84RR18 pKa = 11.84SFLDD22 pKa = 3.1SLLVVLLRR30 pKa = 11.84VVAHH34 pKa = 6.11PVSLVLAVFLVLFVAAEE51 pKa = 4.18VLEE54 pKa = 4.46TTGPLEE60 pKa = 4.97SLDD63 pKa = 3.96KK64 pKa = 11.03LIKK67 pKa = 10.19QEE69 pKa = 5.06LGSKK73 pKa = 10.14DD74 pKa = 3.33INSLEE79 pKa = 3.95KK80 pKa = 10.5FLLKK84 pKa = 10.91GFDD87 pKa = 3.16KK88 pKa = 11.24CIVFVILYY96 pKa = 5.85KK97 pKa = 9.98TKK99 pKa = 10.5VVATLAYY106 pKa = 10.31SIFVALNPTKK116 pKa = 10.59LRR118 pKa = 11.84WSVFGAAVLIVVAIPSLPVFYY139 pKa = 10.59HH140 pKa = 7.08IITAVALVFYY150 pKa = 10.53LALQRR155 pKa = 11.84IEE157 pKa = 4.99HH158 pKa = 6.57KK159 pKa = 10.63ALTVFIYY166 pKa = 9.97VAGMVLYY173 pKa = 9.03TSAVVVSVATAGAGKK188 pKa = 10.14RR189 pKa = 11.84NSTGAPP195 pKa = 3.29
Molecular weight: 21.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3205 |
170 |
2444 |
801.3 |
90.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.27 ± 1.251 | 2.059 ± 0.506 |
6.115 ± 0.857 | 5.367 ± 0.49 |
4.618 ± 0.437 | 5.491 ± 0.668 |
2.559 ± 0.349 | 5.523 ± 0.27 |
5.086 ± 0.317 | 9.735 ± 1.731 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.309 ± 0.463 | 2.964 ± 0.579 |
4.399 ± 0.272 | 3.401 ± 0.622 |
6.739 ± 0.651 | 6.646 ± 0.053 |
5.71 ± 0.611 | 8.924 ± 1.751 |
0.78 ± 0.23 | 4.306 ± 0.271 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
