
Vaprio virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Vaprio ledantevirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
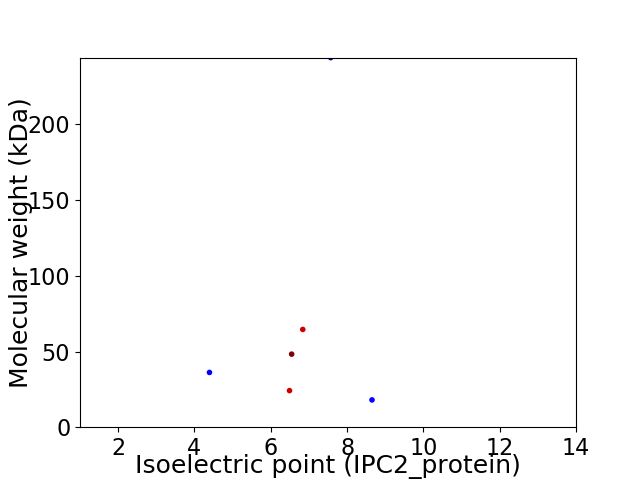
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1BSY6|A0A2P1BSY6_9RHAB Nucleocapsid protein OS=Vaprio virus OX=2100727 PE=4 SV=1
MM1 pKa = 6.94NRR3 pKa = 11.84SRR5 pKa = 11.84VKK7 pKa = 10.51EE8 pKa = 3.56LANRR12 pKa = 11.84QFFKK16 pKa = 10.74DD17 pKa = 3.34IEE19 pKa = 4.41GKK21 pKa = 10.05QFDD24 pKa = 4.17EE25 pKa = 6.16DD26 pKa = 4.15EE27 pKa = 5.69DD28 pKa = 3.9EE29 pKa = 4.72LTYY32 pKa = 10.83QDD34 pKa = 4.15VKK36 pKa = 10.46PTGSNLFEE44 pKa = 4.48RR45 pKa = 11.84KK46 pKa = 9.05EE47 pKa = 4.08DD48 pKa = 3.57EE49 pKa = 4.31LEE51 pKa = 3.88IRR53 pKa = 11.84PEE55 pKa = 3.74EE56 pKa = 4.11EE57 pKa = 3.76EE58 pKa = 3.82FDD60 pKa = 3.81EE61 pKa = 5.01EE62 pKa = 4.16EE63 pKa = 4.48SEE65 pKa = 4.38EE66 pKa = 4.0EE67 pKa = 4.37GEE69 pKa = 5.15GIEE72 pKa = 5.03EE73 pKa = 5.94DD74 pKa = 3.89LDD76 pKa = 3.51QDD78 pKa = 3.83QDD80 pKa = 4.29VEE82 pKa = 4.34FHH84 pKa = 7.29DD85 pKa = 4.99PPEE88 pKa = 4.35QVDD91 pKa = 3.94SEE93 pKa = 4.4NDD95 pKa = 3.44GMVSAVEE102 pKa = 4.01NLDD105 pKa = 3.43EE106 pKa = 4.38EE107 pKa = 4.63SRR109 pKa = 11.84EE110 pKa = 3.76EE111 pKa = 4.17SYY113 pKa = 10.92IEE115 pKa = 4.65SEE117 pKa = 4.33LNHH120 pKa = 6.61LALSDD125 pKa = 3.65HH126 pKa = 6.8LEE128 pKa = 4.32SKK130 pKa = 10.37MSASEE135 pKa = 4.03KK136 pKa = 10.12AASSEE141 pKa = 4.34DD142 pKa = 3.43EE143 pKa = 4.36SEE145 pKa = 3.98PDD147 pKa = 3.09EE148 pKa = 4.87VKK150 pKa = 10.52IEE152 pKa = 3.92FDD154 pKa = 4.25KK155 pKa = 11.25IQLPAVRR162 pKa = 11.84SQEE165 pKa = 4.1EE166 pKa = 3.97LDD168 pKa = 4.78DD169 pKa = 3.56ICFGMFHH176 pKa = 7.01ALVSKK181 pKa = 10.5IGWSLIPLHH190 pKa = 6.72CKK192 pKa = 10.35AGPKK196 pKa = 9.07TLTVSMAPVGKK207 pKa = 9.49PKK209 pKa = 10.25RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84NSSYY216 pKa = 11.13NEE218 pKa = 3.77EE219 pKa = 4.09PPILEE224 pKa = 4.07NLKK227 pKa = 10.01EE228 pKa = 4.17VNKK231 pKa = 10.26GKK233 pKa = 10.44SSEE236 pKa = 4.3TIEE239 pKa = 4.09EE240 pKa = 4.2NKK242 pKa = 10.52DD243 pKa = 3.2GRR245 pKa = 11.84FKK247 pKa = 11.8DD248 pKa = 3.58MVSKK252 pKa = 10.61IRR254 pKa = 11.84AGQIRR259 pKa = 11.84FMKK262 pKa = 10.12KK263 pKa = 9.7KK264 pKa = 9.5NNGYY268 pKa = 8.4MVLGMDD274 pKa = 4.04TPGMSEE280 pKa = 4.73DD281 pKa = 4.45LIEE284 pKa = 4.6RR285 pKa = 11.84CLNNSANEE293 pKa = 4.19GEE295 pKa = 4.6AVEE298 pKa = 5.17KK299 pKa = 10.25MLEE302 pKa = 4.14EE303 pKa = 4.17TGMLPLVQALCKK315 pKa = 10.65YY316 pKa = 9.41PP317 pKa = 3.48
MM1 pKa = 6.94NRR3 pKa = 11.84SRR5 pKa = 11.84VKK7 pKa = 10.51EE8 pKa = 3.56LANRR12 pKa = 11.84QFFKK16 pKa = 10.74DD17 pKa = 3.34IEE19 pKa = 4.41GKK21 pKa = 10.05QFDD24 pKa = 4.17EE25 pKa = 6.16DD26 pKa = 4.15EE27 pKa = 5.69DD28 pKa = 3.9EE29 pKa = 4.72LTYY32 pKa = 10.83QDD34 pKa = 4.15VKK36 pKa = 10.46PTGSNLFEE44 pKa = 4.48RR45 pKa = 11.84KK46 pKa = 9.05EE47 pKa = 4.08DD48 pKa = 3.57EE49 pKa = 4.31LEE51 pKa = 3.88IRR53 pKa = 11.84PEE55 pKa = 3.74EE56 pKa = 4.11EE57 pKa = 3.76EE58 pKa = 3.82FDD60 pKa = 3.81EE61 pKa = 5.01EE62 pKa = 4.16EE63 pKa = 4.48SEE65 pKa = 4.38EE66 pKa = 4.0EE67 pKa = 4.37GEE69 pKa = 5.15GIEE72 pKa = 5.03EE73 pKa = 5.94DD74 pKa = 3.89LDD76 pKa = 3.51QDD78 pKa = 3.83QDD80 pKa = 4.29VEE82 pKa = 4.34FHH84 pKa = 7.29DD85 pKa = 4.99PPEE88 pKa = 4.35QVDD91 pKa = 3.94SEE93 pKa = 4.4NDD95 pKa = 3.44GMVSAVEE102 pKa = 4.01NLDD105 pKa = 3.43EE106 pKa = 4.38EE107 pKa = 4.63SRR109 pKa = 11.84EE110 pKa = 3.76EE111 pKa = 4.17SYY113 pKa = 10.92IEE115 pKa = 4.65SEE117 pKa = 4.33LNHH120 pKa = 6.61LALSDD125 pKa = 3.65HH126 pKa = 6.8LEE128 pKa = 4.32SKK130 pKa = 10.37MSASEE135 pKa = 4.03KK136 pKa = 10.12AASSEE141 pKa = 4.34DD142 pKa = 3.43EE143 pKa = 4.36SEE145 pKa = 3.98PDD147 pKa = 3.09EE148 pKa = 4.87VKK150 pKa = 10.52IEE152 pKa = 3.92FDD154 pKa = 4.25KK155 pKa = 11.25IQLPAVRR162 pKa = 11.84SQEE165 pKa = 4.1EE166 pKa = 3.97LDD168 pKa = 4.78DD169 pKa = 3.56ICFGMFHH176 pKa = 7.01ALVSKK181 pKa = 10.5IGWSLIPLHH190 pKa = 6.72CKK192 pKa = 10.35AGPKK196 pKa = 9.07TLTVSMAPVGKK207 pKa = 9.49PKK209 pKa = 10.25RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84NSSYY216 pKa = 11.13NEE218 pKa = 3.77EE219 pKa = 4.09PPILEE224 pKa = 4.07NLKK227 pKa = 10.01EE228 pKa = 4.17VNKK231 pKa = 10.26GKK233 pKa = 10.44SSEE236 pKa = 4.3TIEE239 pKa = 4.09EE240 pKa = 4.2NKK242 pKa = 10.52DD243 pKa = 3.2GRR245 pKa = 11.84FKK247 pKa = 11.8DD248 pKa = 3.58MVSKK252 pKa = 10.61IRR254 pKa = 11.84AGQIRR259 pKa = 11.84FMKK262 pKa = 10.12KK263 pKa = 9.7KK264 pKa = 9.5NNGYY268 pKa = 8.4MVLGMDD274 pKa = 4.04TPGMSEE280 pKa = 4.73DD281 pKa = 4.45LIEE284 pKa = 4.6RR285 pKa = 11.84CLNNSANEE293 pKa = 4.19GEE295 pKa = 4.6AVEE298 pKa = 5.17KK299 pKa = 10.25MLEE302 pKa = 4.14EE303 pKa = 4.17TGMLPLVQALCKK315 pKa = 10.65YY316 pKa = 9.41PP317 pKa = 3.48
Molecular weight: 36.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1BSX2|A0A2P1BSX2_9RHAB Matrix OS=Vaprio virus OX=2100727 PE=4 SV=1
MM1 pKa = 7.15AQARR5 pKa = 11.84SEE7 pKa = 4.23SGQEE11 pKa = 3.53NQLRR15 pKa = 11.84KK16 pKa = 10.18GRR18 pKa = 11.84TIMSRR23 pKa = 11.84FKK25 pKa = 10.99SSFSSHH31 pKa = 5.8VDD33 pKa = 3.17SGPKK37 pKa = 9.93SVTVLIATSDD47 pKa = 3.58SDD49 pKa = 3.39IKK51 pKa = 10.78IAIGQDD57 pKa = 2.94KK58 pKa = 10.34KK59 pKa = 10.97INEE62 pKa = 4.84LIDD65 pKa = 3.4YY66 pKa = 10.27SVIGKK71 pKa = 8.81DD72 pKa = 3.27EE73 pKa = 4.05KK74 pKa = 10.15RR75 pKa = 11.84TKK77 pKa = 10.28KK78 pKa = 9.54IQRR81 pKa = 11.84ANMVWKK87 pKa = 10.46QNYY90 pKa = 6.23EE91 pKa = 3.99TEE93 pKa = 4.2KK94 pKa = 11.06VNNCLYY100 pKa = 10.11CWKK103 pKa = 10.34GSQGRR108 pKa = 11.84MKK110 pKa = 10.15KK111 pKa = 10.53VSFFEE116 pKa = 4.19SDD118 pKa = 2.94EE119 pKa = 4.55FIYY122 pKa = 10.24VQNEE126 pKa = 3.46QGMMDD131 pKa = 4.46KK132 pKa = 10.79YY133 pKa = 11.28VALNDD138 pKa = 4.01PKK140 pKa = 10.97NLQKK144 pKa = 10.95GKK146 pKa = 8.15EE147 pKa = 4.19VKK149 pKa = 10.32LHH151 pKa = 6.02GPIPEE156 pKa = 4.21YY157 pKa = 11.15
MM1 pKa = 7.15AQARR5 pKa = 11.84SEE7 pKa = 4.23SGQEE11 pKa = 3.53NQLRR15 pKa = 11.84KK16 pKa = 10.18GRR18 pKa = 11.84TIMSRR23 pKa = 11.84FKK25 pKa = 10.99SSFSSHH31 pKa = 5.8VDD33 pKa = 3.17SGPKK37 pKa = 9.93SVTVLIATSDD47 pKa = 3.58SDD49 pKa = 3.39IKK51 pKa = 10.78IAIGQDD57 pKa = 2.94KK58 pKa = 10.34KK59 pKa = 10.97INEE62 pKa = 4.84LIDD65 pKa = 3.4YY66 pKa = 10.27SVIGKK71 pKa = 8.81DD72 pKa = 3.27EE73 pKa = 4.05KK74 pKa = 10.15RR75 pKa = 11.84TKK77 pKa = 10.28KK78 pKa = 9.54IQRR81 pKa = 11.84ANMVWKK87 pKa = 10.46QNYY90 pKa = 6.23EE91 pKa = 3.99TEE93 pKa = 4.2KK94 pKa = 11.06VNNCLYY100 pKa = 10.11CWKK103 pKa = 10.34GSQGRR108 pKa = 11.84MKK110 pKa = 10.15KK111 pKa = 10.53VSFFEE116 pKa = 4.19SDD118 pKa = 2.94EE119 pKa = 4.55FIYY122 pKa = 10.24VQNEE126 pKa = 3.46QGMMDD131 pKa = 4.46KK132 pKa = 10.79YY133 pKa = 11.28VALNDD138 pKa = 4.01PKK140 pKa = 10.97NLQKK144 pKa = 10.95GKK146 pKa = 8.15EE147 pKa = 4.19VKK149 pKa = 10.32LHH151 pKa = 6.02GPIPEE156 pKa = 4.21YY157 pKa = 11.15
Molecular weight: 18.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3807 |
157 |
2127 |
634.5 |
72.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.571 ± 1.046 | 1.839 ± 0.197 |
6.12 ± 0.257 | 7.407 ± 1.359 |
4.597 ± 0.322 | 5.963 ± 0.33 |
2.679 ± 0.31 | 6.436 ± 0.556 |
7.985 ± 0.513 | 9.614 ± 1.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.706 ± 0.278 | 4.833 ± 0.449 |
4.492 ± 0.35 | 2.758 ± 0.423 |
5.096 ± 0.341 | 7.434 ± 0.505 |
4.781 ± 0.443 | 5.779 ± 0.252 |
1.629 ± 0.227 | 3.283 ± 0.305 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
