
Alcube virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Alcube phlebovirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
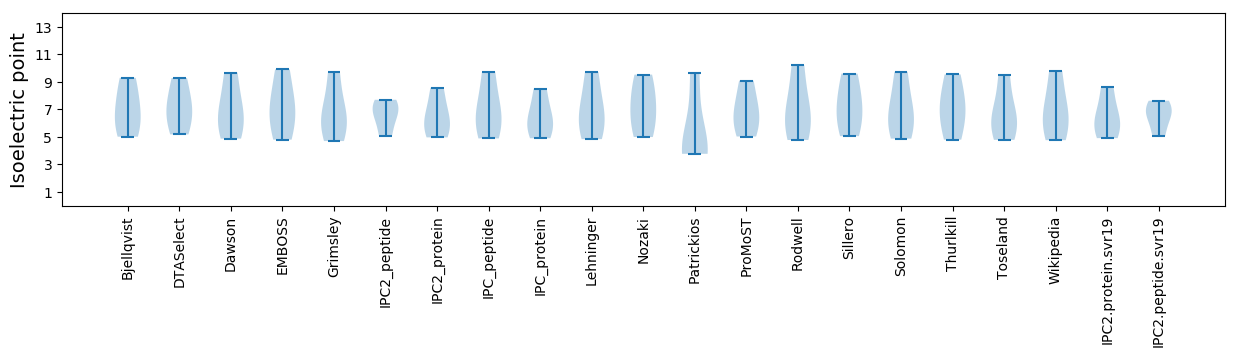
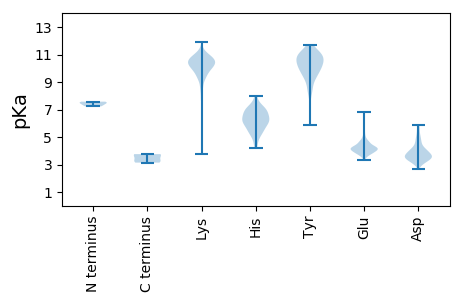
Protein with the lowest isoelectric point:
>tr|A0A0N9MPA8|A0A0N9MPA8_9VIRU Nonstructural protein OS=Alcube virus OX=1725367 PE=4 SV=1
MM1 pKa = 7.39SLHH4 pKa = 5.84YY5 pKa = 10.46LYY7 pKa = 11.13DD8 pKa = 3.91RR9 pKa = 11.84VTPSYY14 pKa = 10.5DD15 pKa = 3.26YY16 pKa = 11.04PNIGEE21 pKa = 4.21VVANFVAVNSNVGLPVCVYY40 pKa = 11.23DD41 pKa = 5.02DD42 pKa = 5.83LEE44 pKa = 5.37FEE46 pKa = 4.6LAGHH50 pKa = 6.12RR51 pKa = 11.84QSLTYY56 pKa = 10.39RR57 pKa = 11.84EE58 pKa = 4.11TLFDD62 pKa = 5.75FIDD65 pKa = 3.93NDD67 pKa = 3.67EE68 pKa = 4.46MPWRR72 pKa = 11.84WGPRR76 pKa = 11.84LWGSRR81 pKa = 11.84VTLDD85 pKa = 3.96EE86 pKa = 4.55VPLLDD91 pKa = 4.45PLMKK95 pKa = 9.89TFQGLPVTDD104 pKa = 4.51LMDD107 pKa = 4.55PRR109 pKa = 11.84LHH111 pKa = 5.96FTRR114 pKa = 11.84NALFWPLDD122 pKa = 3.72HH123 pKa = 7.28VSLKK127 pKa = 10.62AFRR130 pKa = 11.84ILYY133 pKa = 7.18PCRR136 pKa = 11.84FVSFNSSRR144 pKa = 11.84RR145 pKa = 11.84EE146 pKa = 3.43LAEE149 pKa = 4.08YY150 pKa = 10.22IFEE153 pKa = 4.18ATRR156 pKa = 11.84APNLEE161 pKa = 4.17SGLVQLAKK169 pKa = 10.31KK170 pKa = 9.47VRR172 pKa = 11.84DD173 pKa = 3.55MALSLGLTEE182 pKa = 5.57RR183 pKa = 11.84LVPCKK188 pKa = 10.5DD189 pKa = 3.34IIKK192 pKa = 9.9SICILQFIRR201 pKa = 11.84MLKK204 pKa = 10.16GFQLDD209 pKa = 3.65RR210 pKa = 11.84EE211 pKa = 4.1RR212 pKa = 11.84SILQGGTLIPFMIGCTEE229 pKa = 3.71AVVRR233 pKa = 11.84DD234 pKa = 4.27FPGLISTWEE243 pKa = 3.75PRR245 pKa = 11.84LNVVDD250 pKa = 4.86FLHH253 pKa = 5.23VTPYY257 pKa = 11.13DD258 pKa = 3.67SGVSTDD264 pKa = 4.18SEE266 pKa = 4.22FDD268 pKa = 3.16SDD270 pKa = 4.69IEE272 pKa = 4.3VV273 pKa = 3.14
MM1 pKa = 7.39SLHH4 pKa = 5.84YY5 pKa = 10.46LYY7 pKa = 11.13DD8 pKa = 3.91RR9 pKa = 11.84VTPSYY14 pKa = 10.5DD15 pKa = 3.26YY16 pKa = 11.04PNIGEE21 pKa = 4.21VVANFVAVNSNVGLPVCVYY40 pKa = 11.23DD41 pKa = 5.02DD42 pKa = 5.83LEE44 pKa = 5.37FEE46 pKa = 4.6LAGHH50 pKa = 6.12RR51 pKa = 11.84QSLTYY56 pKa = 10.39RR57 pKa = 11.84EE58 pKa = 4.11TLFDD62 pKa = 5.75FIDD65 pKa = 3.93NDD67 pKa = 3.67EE68 pKa = 4.46MPWRR72 pKa = 11.84WGPRR76 pKa = 11.84LWGSRR81 pKa = 11.84VTLDD85 pKa = 3.96EE86 pKa = 4.55VPLLDD91 pKa = 4.45PLMKK95 pKa = 9.89TFQGLPVTDD104 pKa = 4.51LMDD107 pKa = 4.55PRR109 pKa = 11.84LHH111 pKa = 5.96FTRR114 pKa = 11.84NALFWPLDD122 pKa = 3.72HH123 pKa = 7.28VSLKK127 pKa = 10.62AFRR130 pKa = 11.84ILYY133 pKa = 7.18PCRR136 pKa = 11.84FVSFNSSRR144 pKa = 11.84RR145 pKa = 11.84EE146 pKa = 3.43LAEE149 pKa = 4.08YY150 pKa = 10.22IFEE153 pKa = 4.18ATRR156 pKa = 11.84APNLEE161 pKa = 4.17SGLVQLAKK169 pKa = 10.31KK170 pKa = 9.47VRR172 pKa = 11.84DD173 pKa = 3.55MALSLGLTEE182 pKa = 5.57RR183 pKa = 11.84LVPCKK188 pKa = 10.5DD189 pKa = 3.34IIKK192 pKa = 9.9SICILQFIRR201 pKa = 11.84MLKK204 pKa = 10.16GFQLDD209 pKa = 3.65RR210 pKa = 11.84EE211 pKa = 4.1RR212 pKa = 11.84SILQGGTLIPFMIGCTEE229 pKa = 3.71AVVRR233 pKa = 11.84DD234 pKa = 4.27FPGLISTWEE243 pKa = 3.75PRR245 pKa = 11.84LNVVDD250 pKa = 4.86FLHH253 pKa = 5.23VTPYY257 pKa = 11.13DD258 pKa = 3.67SGVSTDD264 pKa = 4.18SEE266 pKa = 4.22FDD268 pKa = 3.16SDD270 pKa = 4.69IEE272 pKa = 4.3VV273 pKa = 3.14
Molecular weight: 31.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N9MPA8|A0A0N9MPA8_9VIRU Nonstructural protein OS=Alcube virus OX=1725367 PE=4 SV=1
MM1 pKa = 7.57SGQEE5 pKa = 4.1EE6 pKa = 4.18FAKK9 pKa = 10.44IAVEE13 pKa = 4.36FASEE17 pKa = 4.11AVDD20 pKa = 3.25TTLILEE26 pKa = 4.27IVKK29 pKa = 9.85EE30 pKa = 4.16FAYY33 pKa = 10.15QGYY36 pKa = 9.31DD37 pKa = 2.91AARR40 pKa = 11.84VIEE43 pKa = 3.95LVRR46 pKa = 11.84EE47 pKa = 4.2KK48 pKa = 11.12GGEE51 pKa = 3.74TWRR54 pKa = 11.84NDD56 pKa = 3.36VKK58 pKa = 10.65TMIIIALTRR67 pKa = 11.84GNKK70 pKa = 7.66PAKK73 pKa = 9.52ILEE76 pKa = 4.29KK77 pKa = 9.96MSVSGKK83 pKa = 7.83TKK85 pKa = 10.11FRR87 pKa = 11.84ALVLKK92 pKa = 10.89YY93 pKa = 10.15GLKK96 pKa = 10.29SGNPGRR102 pKa = 11.84DD103 pKa = 3.46DD104 pKa = 3.39LTLARR109 pKa = 11.84IASAFAAWTIQAIKK123 pKa = 10.41VVEE126 pKa = 4.66AYY128 pKa = 10.89LPVTGAAMDD137 pKa = 3.96EE138 pKa = 4.33LSPGFPRR145 pKa = 11.84PMMHH149 pKa = 6.95PCFGGLIDD157 pKa = 3.73NTLPEE162 pKa = 4.27EE163 pKa = 4.48TVASLVKK170 pKa = 10.49AHH172 pKa = 6.7SLFLDD177 pKa = 3.94AFSRR181 pKa = 11.84TINPSMRR188 pKa = 11.84TKK190 pKa = 10.44TKK192 pKa = 11.08AEE194 pKa = 3.9VAKK197 pKa = 10.96SFEE200 pKa = 4.22QPLNAAINSRR210 pKa = 11.84FLTSDD215 pKa = 3.59AKK217 pKa = 10.78RR218 pKa = 11.84KK219 pKa = 8.56ILKK222 pKa = 9.71SAGVIDD228 pKa = 4.63TNLKK232 pKa = 8.82PAPAVEE238 pKa = 4.09MAAKK242 pKa = 10.05KK243 pKa = 9.94FSEE246 pKa = 4.45MAA248 pKa = 3.73
MM1 pKa = 7.57SGQEE5 pKa = 4.1EE6 pKa = 4.18FAKK9 pKa = 10.44IAVEE13 pKa = 4.36FASEE17 pKa = 4.11AVDD20 pKa = 3.25TTLILEE26 pKa = 4.27IVKK29 pKa = 9.85EE30 pKa = 4.16FAYY33 pKa = 10.15QGYY36 pKa = 9.31DD37 pKa = 2.91AARR40 pKa = 11.84VIEE43 pKa = 3.95LVRR46 pKa = 11.84EE47 pKa = 4.2KK48 pKa = 11.12GGEE51 pKa = 3.74TWRR54 pKa = 11.84NDD56 pKa = 3.36VKK58 pKa = 10.65TMIIIALTRR67 pKa = 11.84GNKK70 pKa = 7.66PAKK73 pKa = 9.52ILEE76 pKa = 4.29KK77 pKa = 9.96MSVSGKK83 pKa = 7.83TKK85 pKa = 10.11FRR87 pKa = 11.84ALVLKK92 pKa = 10.89YY93 pKa = 10.15GLKK96 pKa = 10.29SGNPGRR102 pKa = 11.84DD103 pKa = 3.46DD104 pKa = 3.39LTLARR109 pKa = 11.84IASAFAAWTIQAIKK123 pKa = 10.41VVEE126 pKa = 4.66AYY128 pKa = 10.89LPVTGAAMDD137 pKa = 3.96EE138 pKa = 4.33LSPGFPRR145 pKa = 11.84PMMHH149 pKa = 6.95PCFGGLIDD157 pKa = 3.73NTLPEE162 pKa = 4.27EE163 pKa = 4.48TVASLVKK170 pKa = 10.49AHH172 pKa = 6.7SLFLDD177 pKa = 3.94AFSRR181 pKa = 11.84TINPSMRR188 pKa = 11.84TKK190 pKa = 10.44TKK192 pKa = 11.08AEE194 pKa = 3.9VAKK197 pKa = 10.96SFEE200 pKa = 4.22QPLNAAINSRR210 pKa = 11.84FLTSDD215 pKa = 3.59AKK217 pKa = 10.78RR218 pKa = 11.84KK219 pKa = 8.56ILKK222 pKa = 9.71SAGVIDD228 pKa = 4.63TNLKK232 pKa = 8.82PAPAVEE238 pKa = 4.09MAAKK242 pKa = 10.05KK243 pKa = 9.94FSEE246 pKa = 4.45MAA248 pKa = 3.73
Molecular weight: 27.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3948 |
248 |
2096 |
987.0 |
110.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.003 ± 1.234 | 2.66 ± 0.918 |
5.699 ± 0.394 | 6.307 ± 0.532 |
4.357 ± 0.448 | 5.927 ± 0.358 |
2.482 ± 0.375 | 6.104 ± 0.119 |
5.8 ± 0.529 | 9.271 ± 0.467 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.267 ± 0.527 | 3.647 ± 0.306 |
4.179 ± 0.391 | 2.913 ± 0.316 |
5.496 ± 0.59 | 9.093 ± 0.766 |
5.674 ± 0.247 | 7.042 ± 0.525 |
1.292 ± 0.129 | 2.786 ± 0.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
