
Torque teno midi virus 6
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
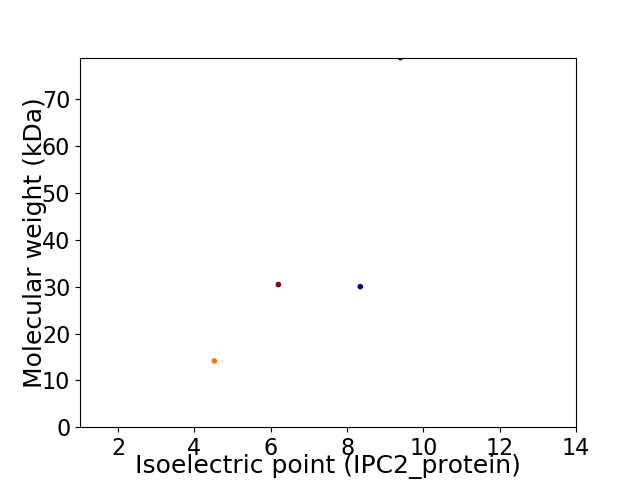
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A7VLW0|A7VLW0_9VIRU Capsid protein OS=Torque teno midi virus 6 OX=2065047 PE=3 SV=1
MM1 pKa = 7.13QNISAEE7 pKa = 4.14DD8 pKa = 4.34FYY10 pKa = 11.74KK11 pKa = 9.89EE12 pKa = 3.77THH14 pKa = 5.91FNQVTKK20 pKa = 10.09HH21 pKa = 4.95QLWMSIVTDD30 pKa = 3.48AHH32 pKa = 7.37DD33 pKa = 3.92SVCNCWHH40 pKa = 7.21PFAHH44 pKa = 6.13MLANIFPPGHH54 pKa = 7.2KK55 pKa = 10.11DD56 pKa = 3.09RR57 pKa = 11.84NLTINQILEE66 pKa = 4.12RR67 pKa = 11.84DD68 pKa = 3.96LKK70 pKa = 9.31EE71 pKa = 3.99TWPSGGDD78 pKa = 3.01ADD80 pKa = 4.49ASHH83 pKa = 6.68GLVAGEE89 pKa = 4.25DD90 pKa = 3.38TGGEE94 pKa = 3.91QIINPEE100 pKa = 4.16EE101 pKa = 4.38KK102 pKa = 9.96EE103 pKa = 3.83EE104 pKa = 4.08EE105 pKa = 4.31YY106 pKa = 10.57IEE108 pKa = 5.11DD109 pKa = 3.84EE110 pKa = 5.11EE111 pKa = 4.32ITEE114 pKa = 4.26LLKK117 pKa = 11.03AADD120 pKa = 3.72VAATRR125 pKa = 4.11
MM1 pKa = 7.13QNISAEE7 pKa = 4.14DD8 pKa = 4.34FYY10 pKa = 11.74KK11 pKa = 9.89EE12 pKa = 3.77THH14 pKa = 5.91FNQVTKK20 pKa = 10.09HH21 pKa = 4.95QLWMSIVTDD30 pKa = 3.48AHH32 pKa = 7.37DD33 pKa = 3.92SVCNCWHH40 pKa = 7.21PFAHH44 pKa = 6.13MLANIFPPGHH54 pKa = 7.2KK55 pKa = 10.11DD56 pKa = 3.09RR57 pKa = 11.84NLTINQILEE66 pKa = 4.12RR67 pKa = 11.84DD68 pKa = 3.96LKK70 pKa = 9.31EE71 pKa = 3.99TWPSGGDD78 pKa = 3.01ADD80 pKa = 4.49ASHH83 pKa = 6.68GLVAGEE89 pKa = 4.25DD90 pKa = 3.38TGGEE94 pKa = 3.91QIINPEE100 pKa = 4.16EE101 pKa = 4.38KK102 pKa = 9.96EE103 pKa = 3.83EE104 pKa = 4.08EE105 pKa = 4.31YY106 pKa = 10.57IEE108 pKa = 5.11DD109 pKa = 3.84EE110 pKa = 5.11EE111 pKa = 4.32ITEE114 pKa = 4.26LLKK117 pKa = 11.03AADD120 pKa = 3.72VAATRR125 pKa = 4.11
Molecular weight: 14.15 kDa
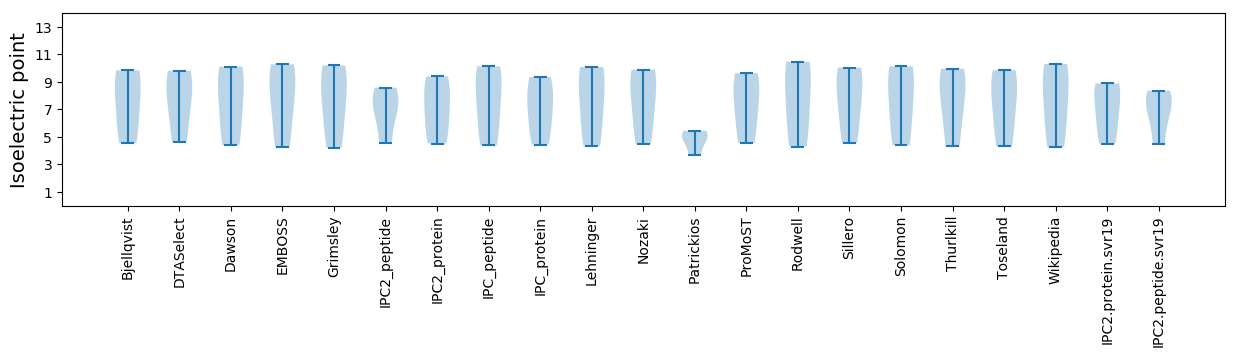
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLW0|A7VLW0_9VIRU Capsid protein OS=Torque teno midi virus 6 OX=2065047 PE=3 SV=1
MM1 pKa = 7.28AFWWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.65PWFGRR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.61RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84TNYY25 pKa = 9.37KK26 pKa = 8.32PRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.42RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84IYY35 pKa = 10.01RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84NYY41 pKa = 8.44RR42 pKa = 11.84TTKK45 pKa = 8.14GRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84YY52 pKa = 8.58KK53 pKa = 10.44VRR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 6.7QQKK60 pKa = 9.41ILVKK64 pKa = 9.89QWQPEE69 pKa = 4.43CIRR72 pKa = 11.84KK73 pKa = 9.54CKK75 pKa = 10.19IKK77 pKa = 10.83GLGTIVLGAEE87 pKa = 4.3GTQYY91 pKa = 10.98RR92 pKa = 11.84CYY94 pKa = 10.45TDD96 pKa = 5.86YY97 pKa = 11.43LDD99 pKa = 3.85EE100 pKa = 4.2WTNPKK105 pKa = 10.05VPGGGGFGCEE115 pKa = 4.06LFTLRR120 pKa = 11.84YY121 pKa = 8.47LYY123 pKa = 11.14KK124 pKa = 10.04MYY126 pKa = 10.65LARR129 pKa = 11.84KK130 pKa = 8.94NIWTFSNKK138 pKa = 9.81YY139 pKa = 9.31KK140 pKa = 10.9DD141 pKa = 3.52LVRR144 pKa = 11.84YY145 pKa = 7.41TGTRR149 pKa = 11.84FTFFRR154 pKa = 11.84HH155 pKa = 6.31PEE157 pKa = 3.7TDD159 pKa = 4.1FILAYY164 pKa = 10.21DD165 pKa = 4.06IQPPFHH171 pKa = 5.84ITKK174 pKa = 6.88WTYY177 pKa = 10.62MSIHH181 pKa = 6.44PQMLLLRR188 pKa = 11.84KK189 pKa = 9.18HH190 pKa = 6.28KK191 pKa = 10.52KK192 pKa = 10.08LLLSTATKK200 pKa = 10.5PNGKK204 pKa = 8.62LTTRR208 pKa = 11.84FKK210 pKa = 10.59IRR212 pKa = 11.84PPKK215 pKa = 10.19QLVTKK220 pKa = 9.8WFFQKK225 pKa = 10.77DD226 pKa = 3.34FAQYY230 pKa = 11.02GLVAIAAAACNLRR243 pKa = 11.84YY244 pKa = 8.88PWLACCNEE252 pKa = 3.76NLIITLRR259 pKa = 11.84YY260 pKa = 8.61LQPQFYY266 pKa = 10.15HH267 pKa = 6.29NSAWAQTTKK276 pKa = 10.51EE277 pKa = 4.02YY278 pKa = 11.15NPVYY282 pKa = 9.47GTQPMKK288 pKa = 10.16KK289 pKa = 7.61TLTFNYY295 pKa = 9.85IIGNQRR301 pKa = 11.84GTYY304 pKa = 8.84TMSNSDD310 pKa = 2.83ISDD313 pKa = 3.55YY314 pKa = 11.21DD315 pKa = 3.84SSVSISKK322 pKa = 9.91GWFQPKK328 pKa = 9.4ILNAYY333 pKa = 6.6EE334 pKa = 4.4VKK336 pKa = 10.85DD337 pKa = 3.7NTVTQALTPCGVLRR351 pKa = 11.84YY352 pKa = 10.05NPADD356 pKa = 5.09DD357 pKa = 5.84DD358 pKa = 4.4GQGNKK363 pKa = 9.21LWVVNTLTYY372 pKa = 9.76NWKK375 pKa = 10.17IPTDD379 pKa = 3.83DD380 pKa = 4.34DD381 pKa = 5.51LILEE385 pKa = 5.53GYY387 pKa = 8.49PLWLMLYY394 pKa = 9.85GYY396 pKa = 9.17TSFLYY401 pKa = 9.84QVKK404 pKa = 10.29ADD406 pKa = 4.07KK407 pKa = 11.4NPFSSSLIVVKK418 pKa = 10.6SSALQRR424 pKa = 11.84VFGTDD429 pKa = 2.9TTGWYY434 pKa = 9.53PLIDD438 pKa = 3.66KK439 pKa = 10.98SFMDD443 pKa = 4.21GKK445 pKa = 11.22GPGNTDD451 pKa = 4.17PILLHH456 pKa = 6.26RR457 pKa = 11.84KK458 pKa = 7.66YY459 pKa = 9.57WYY461 pKa = 9.27PSLYY465 pKa = 9.75GQRR468 pKa = 11.84DD469 pKa = 4.12SISQLVNCGPYY480 pKa = 8.62VPKK483 pKa = 11.01YY484 pKa = 10.85NEE486 pKa = 4.45TKK488 pKa = 10.85NSTWQCNYY496 pKa = 10.43KK497 pKa = 9.02YY498 pKa = 9.33TFYY501 pKa = 10.82FKK503 pKa = 10.18WGGHH507 pKa = 5.01YY508 pKa = 10.29PPEE511 pKa = 4.12QEE513 pKa = 5.03AEE515 pKa = 4.05NPEE518 pKa = 4.15KK519 pKa = 10.98KK520 pKa = 10.27NIYY523 pKa = 8.91PVPDD527 pKa = 3.6KK528 pKa = 10.81QQEE531 pKa = 4.51TISIADD537 pKa = 4.7PISEE541 pKa = 4.5KK542 pKa = 10.52YY543 pKa = 9.42DD544 pKa = 3.41TVFRR548 pKa = 11.84SWDD551 pKa = 3.37YY552 pKa = 11.36RR553 pKa = 11.84RR554 pKa = 11.84GSLTRR559 pKa = 11.84TAIKK563 pKa = 10.34RR564 pKa = 11.84MQEE567 pKa = 3.77NLEE570 pKa = 4.11SDD572 pKa = 3.73EE573 pKa = 4.51SLSTDD578 pKa = 3.66SAGCSSKK585 pKa = 10.67KK586 pKa = 9.21KK587 pKa = 10.34KK588 pKa = 10.22KK589 pKa = 10.2YY590 pKa = 10.65LPTLQDD596 pKa = 3.22PKK598 pKa = 10.97KK599 pKa = 9.97EE600 pKa = 3.88NKK602 pKa = 9.64EE603 pKa = 3.77IQKK606 pKa = 10.43CLLSLCEE613 pKa = 3.99EE614 pKa = 4.51NTWQEE619 pKa = 4.02PPQDD623 pKa = 4.09SNILQLLQQQHH634 pKa = 5.14QQQQSIKK641 pKa = 10.19QNLLTLIADD650 pKa = 4.66LKK652 pKa = 10.7AKK654 pKa = 9.63QRR656 pKa = 11.84NLLHH660 pKa = 4.72QTGYY664 pKa = 10.74LAA666 pKa = 5.21
MM1 pKa = 7.28AFWWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.65PWFGRR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.61RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84TNYY25 pKa = 9.37KK26 pKa = 8.32PRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.42RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84IYY35 pKa = 10.01RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84NYY41 pKa = 8.44RR42 pKa = 11.84TTKK45 pKa = 8.14GRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84YY52 pKa = 8.58KK53 pKa = 10.44VRR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 6.7QQKK60 pKa = 9.41ILVKK64 pKa = 9.89QWQPEE69 pKa = 4.43CIRR72 pKa = 11.84KK73 pKa = 9.54CKK75 pKa = 10.19IKK77 pKa = 10.83GLGTIVLGAEE87 pKa = 4.3GTQYY91 pKa = 10.98RR92 pKa = 11.84CYY94 pKa = 10.45TDD96 pKa = 5.86YY97 pKa = 11.43LDD99 pKa = 3.85EE100 pKa = 4.2WTNPKK105 pKa = 10.05VPGGGGFGCEE115 pKa = 4.06LFTLRR120 pKa = 11.84YY121 pKa = 8.47LYY123 pKa = 11.14KK124 pKa = 10.04MYY126 pKa = 10.65LARR129 pKa = 11.84KK130 pKa = 8.94NIWTFSNKK138 pKa = 9.81YY139 pKa = 9.31KK140 pKa = 10.9DD141 pKa = 3.52LVRR144 pKa = 11.84YY145 pKa = 7.41TGTRR149 pKa = 11.84FTFFRR154 pKa = 11.84HH155 pKa = 6.31PEE157 pKa = 3.7TDD159 pKa = 4.1FILAYY164 pKa = 10.21DD165 pKa = 4.06IQPPFHH171 pKa = 5.84ITKK174 pKa = 6.88WTYY177 pKa = 10.62MSIHH181 pKa = 6.44PQMLLLRR188 pKa = 11.84KK189 pKa = 9.18HH190 pKa = 6.28KK191 pKa = 10.52KK192 pKa = 10.08LLLSTATKK200 pKa = 10.5PNGKK204 pKa = 8.62LTTRR208 pKa = 11.84FKK210 pKa = 10.59IRR212 pKa = 11.84PPKK215 pKa = 10.19QLVTKK220 pKa = 9.8WFFQKK225 pKa = 10.77DD226 pKa = 3.34FAQYY230 pKa = 11.02GLVAIAAAACNLRR243 pKa = 11.84YY244 pKa = 8.88PWLACCNEE252 pKa = 3.76NLIITLRR259 pKa = 11.84YY260 pKa = 8.61LQPQFYY266 pKa = 10.15HH267 pKa = 6.29NSAWAQTTKK276 pKa = 10.51EE277 pKa = 4.02YY278 pKa = 11.15NPVYY282 pKa = 9.47GTQPMKK288 pKa = 10.16KK289 pKa = 7.61TLTFNYY295 pKa = 9.85IIGNQRR301 pKa = 11.84GTYY304 pKa = 8.84TMSNSDD310 pKa = 2.83ISDD313 pKa = 3.55YY314 pKa = 11.21DD315 pKa = 3.84SSVSISKK322 pKa = 9.91GWFQPKK328 pKa = 9.4ILNAYY333 pKa = 6.6EE334 pKa = 4.4VKK336 pKa = 10.85DD337 pKa = 3.7NTVTQALTPCGVLRR351 pKa = 11.84YY352 pKa = 10.05NPADD356 pKa = 5.09DD357 pKa = 5.84DD358 pKa = 4.4GQGNKK363 pKa = 9.21LWVVNTLTYY372 pKa = 9.76NWKK375 pKa = 10.17IPTDD379 pKa = 3.83DD380 pKa = 4.34DD381 pKa = 5.51LILEE385 pKa = 5.53GYY387 pKa = 8.49PLWLMLYY394 pKa = 9.85GYY396 pKa = 9.17TSFLYY401 pKa = 9.84QVKK404 pKa = 10.29ADD406 pKa = 4.07KK407 pKa = 11.4NPFSSSLIVVKK418 pKa = 10.6SSALQRR424 pKa = 11.84VFGTDD429 pKa = 2.9TTGWYY434 pKa = 9.53PLIDD438 pKa = 3.66KK439 pKa = 10.98SFMDD443 pKa = 4.21GKK445 pKa = 11.22GPGNTDD451 pKa = 4.17PILLHH456 pKa = 6.26RR457 pKa = 11.84KK458 pKa = 7.66YY459 pKa = 9.57WYY461 pKa = 9.27PSLYY465 pKa = 9.75GQRR468 pKa = 11.84DD469 pKa = 4.12SISQLVNCGPYY480 pKa = 8.62VPKK483 pKa = 11.01YY484 pKa = 10.85NEE486 pKa = 4.45TKK488 pKa = 10.85NSTWQCNYY496 pKa = 10.43KK497 pKa = 9.02YY498 pKa = 9.33TFYY501 pKa = 10.82FKK503 pKa = 10.18WGGHH507 pKa = 5.01YY508 pKa = 10.29PPEE511 pKa = 4.12QEE513 pKa = 5.03AEE515 pKa = 4.05NPEE518 pKa = 4.15KK519 pKa = 10.98KK520 pKa = 10.27NIYY523 pKa = 8.91PVPDD527 pKa = 3.6KK528 pKa = 10.81QQEE531 pKa = 4.51TISIADD537 pKa = 4.7PISEE541 pKa = 4.5KK542 pKa = 10.52YY543 pKa = 9.42DD544 pKa = 3.41TVFRR548 pKa = 11.84SWDD551 pKa = 3.37YY552 pKa = 11.36RR553 pKa = 11.84RR554 pKa = 11.84GSLTRR559 pKa = 11.84TAIKK563 pKa = 10.34RR564 pKa = 11.84MQEE567 pKa = 3.77NLEE570 pKa = 4.11SDD572 pKa = 3.73EE573 pKa = 4.51SLSTDD578 pKa = 3.66SAGCSSKK585 pKa = 10.67KK586 pKa = 9.21KK587 pKa = 10.34KK588 pKa = 10.22KK589 pKa = 10.2YY590 pKa = 10.65LPTLQDD596 pKa = 3.22PKK598 pKa = 10.97KK599 pKa = 9.97EE600 pKa = 3.88NKK602 pKa = 9.64EE603 pKa = 3.77IQKK606 pKa = 10.43CLLSLCEE613 pKa = 3.99EE614 pKa = 4.51NTWQEE619 pKa = 4.02PPQDD623 pKa = 4.09SNILQLLQQQHH634 pKa = 5.14QQQQSIKK641 pKa = 10.19QNLLTLIADD650 pKa = 4.66LKK652 pKa = 10.7AKK654 pKa = 9.63QRR656 pKa = 11.84NLLHH660 pKa = 4.72QTGYY664 pKa = 10.74LAA666 pKa = 5.21
Molecular weight: 78.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322 |
125 |
666 |
330.5 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.976 ± 1.478 | 1.664 ± 0.212 |
4.841 ± 0.463 | 6.43 ± 1.872 |
3.555 ± 0.649 | 4.614 ± 0.585 |
3.026 ± 1.131 | 5.144 ± 0.389 |
8.396 ± 0.768 | 8.018 ± 0.77 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.74 ± 0.257 | 5.144 ± 0.098 |
5.976 ± 1.087 | 5.673 ± 0.774 |
6.278 ± 1.07 | 6.051 ± 1.657 |
7.564 ± 0.376 | 3.177 ± 0.208 |
2.345 ± 0.609 | 4.387 ± 1.687 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
