
Sulfurovum sp. AR
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Sulfurovaceae; Sulfurovum; unclassified Sulfurovum
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
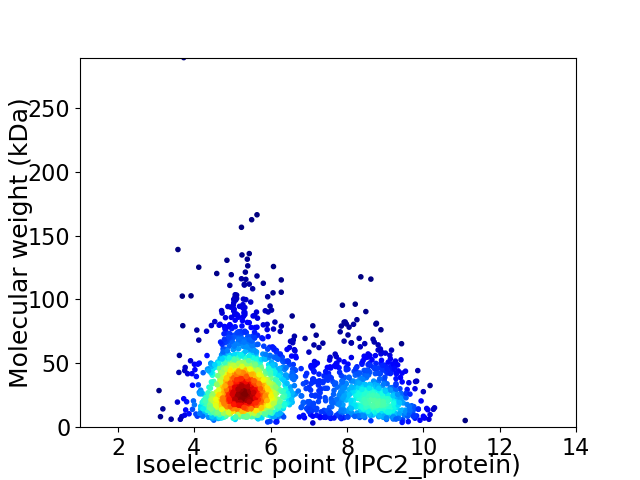
Virtual 2D-PAGE plot for 2114 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I2K6X7|I2K6X7_9PROT Uncharacterized protein OS=Sulfurovum sp. AR OX=1165841 GN=SULAR_06408 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.29LIKK5 pKa = 10.5LIMILGALLLTNLSAAVISGYY26 pKa = 10.31CFEE29 pKa = 5.39DD30 pKa = 3.69VNGDD34 pKa = 3.92GIKK37 pKa = 10.82DD38 pKa = 3.5PGEE41 pKa = 4.02ICAEE45 pKa = 3.88EE46 pKa = 4.19AVWAKK51 pKa = 10.09IVNLSTGHH59 pKa = 6.18VEE61 pKa = 4.01VSKK64 pKa = 10.88NLYY67 pKa = 10.62SIDD70 pKa = 4.41PEE72 pKa = 4.08TSGYY76 pKa = 10.92FEE78 pKa = 5.87FNTHH82 pKa = 5.68SKK84 pKa = 8.81TGQFKK89 pKa = 11.05VFLDD93 pKa = 3.73NNADD97 pKa = 3.73GKK99 pKa = 9.96DD100 pKa = 3.66TVATPPVNTHH110 pKa = 5.58FTVDD114 pKa = 3.68PLGDD118 pKa = 3.63GTDD121 pKa = 3.56TNRR124 pKa = 11.84TNGEE128 pKa = 3.93TYY130 pKa = 8.89YY131 pKa = 10.69TIDD134 pKa = 3.99PGEE137 pKa = 4.25EE138 pKa = 3.56DD139 pKa = 3.96TIYY142 pKa = 10.77EE143 pKa = 4.06NQDD146 pKa = 2.67FGFQFGGDD154 pKa = 4.22CVCEE158 pKa = 4.1GADD161 pKa = 4.3GIIIKK166 pKa = 8.78KK167 pKa = 8.16TININGDD174 pKa = 3.1VSDD177 pKa = 4.25WEE179 pKa = 4.6SVHH182 pKa = 6.73TDD184 pKa = 2.9ADD186 pKa = 4.04NNVCDD191 pKa = 5.06HH192 pKa = 7.03GDD194 pKa = 3.86SEE196 pKa = 5.02DD197 pKa = 3.61KK198 pKa = 11.06DD199 pKa = 5.41AIVQSTGRR207 pKa = 11.84NLVQFTWTGDD217 pKa = 2.91NDD219 pKa = 3.32FAYY222 pKa = 10.36GFTRR226 pKa = 11.84RR227 pKa = 11.84VGSSTNTQTFIYY239 pKa = 10.12YY240 pKa = 10.33ADD242 pKa = 4.1RR243 pKa = 11.84DD244 pKa = 3.48GDD246 pKa = 3.55GRR248 pKa = 11.84MEE250 pKa = 4.81AEE252 pKa = 4.71DD253 pKa = 3.82FALVAGWQGNTGATSLYY270 pKa = 9.51YY271 pKa = 10.45YY272 pKa = 9.88PYY274 pKa = 10.89LPADD278 pKa = 4.25LVNGDD283 pKa = 3.9SMVWTQADD291 pKa = 3.06IDD293 pKa = 4.09AGKK296 pKa = 10.05LPPGGQTPDD305 pKa = 3.43TSWIGSGDD313 pKa = 3.92GYY315 pKa = 9.73TLAGNLDD322 pKa = 3.68TGSGVDD328 pKa = 5.88LIAAGNGLAQGVTEE342 pKa = 4.55ASGGLDD348 pKa = 3.34DD349 pKa = 6.31AGVTMEE355 pKa = 4.37WRR357 pKa = 11.84VRR359 pKa = 11.84WDD361 pKa = 4.02AIGLTPFQGITYY373 pKa = 9.46HH374 pKa = 6.5ISTMNSSVNDD384 pKa = 3.79NNPPGQVDD392 pKa = 4.23DD393 pKa = 5.24NMAGCYY399 pKa = 10.36GEE401 pKa = 4.12ATLRR405 pKa = 11.84YY406 pKa = 9.38CGVTLVPDD414 pKa = 3.99RR415 pKa = 11.84SIILPSGSVGPVYY428 pKa = 10.59FEE430 pKa = 5.51HH431 pKa = 7.24NLTNTGNGDD440 pKa = 3.6EE441 pKa = 4.66NLTLSLTQTEE451 pKa = 4.31SDD453 pKa = 3.66FTVTYY458 pKa = 10.23IEE460 pKa = 4.89YY461 pKa = 11.08YY462 pKa = 10.88NDD464 pKa = 3.51ANGNGVEE471 pKa = 5.02DD472 pKa = 4.09LGEE475 pKa = 4.26EE476 pKa = 4.22NLTGGPLALGAGEE489 pKa = 4.84SINLLVKK496 pKa = 10.55IGLPTGLDD504 pKa = 3.54DD505 pKa = 6.02NIATGEE511 pKa = 4.12INATTTSCAVNKK523 pKa = 10.43SAVVYY528 pKa = 9.82DD529 pKa = 4.11QIRR532 pKa = 11.84VLPLNFDD539 pKa = 3.89LDD541 pKa = 3.64ITKK544 pKa = 10.3EE545 pKa = 4.08VSDD548 pKa = 5.72DD549 pKa = 3.45NASWVDD555 pKa = 3.83TIEE558 pKa = 4.29LSEE561 pKa = 4.51GSPAYY566 pKa = 10.7YY567 pKa = 10.44RR568 pKa = 11.84ITVTNNGPDD577 pKa = 3.32GATDD581 pKa = 3.36VTVNDD586 pKa = 3.7VLPANVNYY594 pKa = 9.86IGYY597 pKa = 9.28YY598 pKa = 8.26ATQGTYY604 pKa = 10.78GAGQWMVGNLALDD617 pKa = 4.42GSAEE621 pKa = 4.01LFIDD625 pKa = 3.85VNVSQTAGDD634 pKa = 3.58LTVIQNIASTTDD646 pKa = 3.19TNDD649 pKa = 3.05TDD651 pKa = 5.16TNDD654 pKa = 3.43QDD656 pKa = 4.34DD657 pKa = 4.36ANITAIHH664 pKa = 6.26IPVVNLQIVKK674 pKa = 8.8TVSNPTPYY682 pKa = 10.31EE683 pKa = 4.25GEE685 pKa = 4.51DD686 pKa = 3.05INYY689 pKa = 7.68TITVTKK695 pKa = 10.49FGIDD699 pKa = 3.31DD700 pKa = 4.01AEE702 pKa = 3.99NVVVNDD708 pKa = 4.34ILPPWVSYY716 pKa = 11.18VDD718 pKa = 3.59STASVGSYY726 pKa = 11.14DD727 pKa = 3.92EE728 pKa = 4.44TTGIWDD734 pKa = 3.77IGTMSNSVEE743 pKa = 4.08TLIIQATVDD752 pKa = 3.61TNATLHH758 pKa = 5.79NPIVNTASVSTDD770 pKa = 3.17TTDD773 pKa = 3.16TNPDD777 pKa = 3.59DD778 pKa = 6.13DD779 pKa = 4.07IASATFNASNPADD792 pKa = 3.86LGITKK797 pKa = 9.07TVDD800 pKa = 3.12NPNPQEE806 pKa = 4.08GDD808 pKa = 3.67TIVYY812 pKa = 7.58TISVTNYY819 pKa = 9.91GPLDD823 pKa = 3.44ATNIYY828 pKa = 9.02VQEE831 pKa = 4.16EE832 pKa = 4.43PFDD835 pKa = 4.44GNLSITGTDD844 pKa = 3.22TTLGTFNAPWWHH856 pKa = 5.91IPEE859 pKa = 4.96LNVSQTAVLTVTAMVDD875 pKa = 3.63LNSTGDD881 pKa = 3.47ILINNVIIDD890 pKa = 4.09TEE892 pKa = 4.11RR893 pKa = 11.84LDD895 pKa = 4.22QNDD898 pKa = 3.99TNGEE902 pKa = 3.93NDD904 pKa = 3.27KK905 pKa = 10.18ATAIVIVGCPCDD917 pKa = 4.58NISSDD922 pKa = 4.11SASAMNKK929 pKa = 8.49TVGLLMICMTLLIGLIFVRR948 pKa = 11.84RR949 pKa = 11.84EE950 pKa = 3.34EE951 pKa = 4.06KK952 pKa = 10.45YY953 pKa = 10.82NRR955 pKa = 11.84NEE957 pKa = 3.7RR958 pKa = 3.68
MM1 pKa = 7.47KK2 pKa = 10.29LIKK5 pKa = 10.5LIMILGALLLTNLSAAVISGYY26 pKa = 10.31CFEE29 pKa = 5.39DD30 pKa = 3.69VNGDD34 pKa = 3.92GIKK37 pKa = 10.82DD38 pKa = 3.5PGEE41 pKa = 4.02ICAEE45 pKa = 3.88EE46 pKa = 4.19AVWAKK51 pKa = 10.09IVNLSTGHH59 pKa = 6.18VEE61 pKa = 4.01VSKK64 pKa = 10.88NLYY67 pKa = 10.62SIDD70 pKa = 4.41PEE72 pKa = 4.08TSGYY76 pKa = 10.92FEE78 pKa = 5.87FNTHH82 pKa = 5.68SKK84 pKa = 8.81TGQFKK89 pKa = 11.05VFLDD93 pKa = 3.73NNADD97 pKa = 3.73GKK99 pKa = 9.96DD100 pKa = 3.66TVATPPVNTHH110 pKa = 5.58FTVDD114 pKa = 3.68PLGDD118 pKa = 3.63GTDD121 pKa = 3.56TNRR124 pKa = 11.84TNGEE128 pKa = 3.93TYY130 pKa = 8.89YY131 pKa = 10.69TIDD134 pKa = 3.99PGEE137 pKa = 4.25EE138 pKa = 3.56DD139 pKa = 3.96TIYY142 pKa = 10.77EE143 pKa = 4.06NQDD146 pKa = 2.67FGFQFGGDD154 pKa = 4.22CVCEE158 pKa = 4.1GADD161 pKa = 4.3GIIIKK166 pKa = 8.78KK167 pKa = 8.16TININGDD174 pKa = 3.1VSDD177 pKa = 4.25WEE179 pKa = 4.6SVHH182 pKa = 6.73TDD184 pKa = 2.9ADD186 pKa = 4.04NNVCDD191 pKa = 5.06HH192 pKa = 7.03GDD194 pKa = 3.86SEE196 pKa = 5.02DD197 pKa = 3.61KK198 pKa = 11.06DD199 pKa = 5.41AIVQSTGRR207 pKa = 11.84NLVQFTWTGDD217 pKa = 2.91NDD219 pKa = 3.32FAYY222 pKa = 10.36GFTRR226 pKa = 11.84RR227 pKa = 11.84VGSSTNTQTFIYY239 pKa = 10.12YY240 pKa = 10.33ADD242 pKa = 4.1RR243 pKa = 11.84DD244 pKa = 3.48GDD246 pKa = 3.55GRR248 pKa = 11.84MEE250 pKa = 4.81AEE252 pKa = 4.71DD253 pKa = 3.82FALVAGWQGNTGATSLYY270 pKa = 9.51YY271 pKa = 10.45YY272 pKa = 9.88PYY274 pKa = 10.89LPADD278 pKa = 4.25LVNGDD283 pKa = 3.9SMVWTQADD291 pKa = 3.06IDD293 pKa = 4.09AGKK296 pKa = 10.05LPPGGQTPDD305 pKa = 3.43TSWIGSGDD313 pKa = 3.92GYY315 pKa = 9.73TLAGNLDD322 pKa = 3.68TGSGVDD328 pKa = 5.88LIAAGNGLAQGVTEE342 pKa = 4.55ASGGLDD348 pKa = 3.34DD349 pKa = 6.31AGVTMEE355 pKa = 4.37WRR357 pKa = 11.84VRR359 pKa = 11.84WDD361 pKa = 4.02AIGLTPFQGITYY373 pKa = 9.46HH374 pKa = 6.5ISTMNSSVNDD384 pKa = 3.79NNPPGQVDD392 pKa = 4.23DD393 pKa = 5.24NMAGCYY399 pKa = 10.36GEE401 pKa = 4.12ATLRR405 pKa = 11.84YY406 pKa = 9.38CGVTLVPDD414 pKa = 3.99RR415 pKa = 11.84SIILPSGSVGPVYY428 pKa = 10.59FEE430 pKa = 5.51HH431 pKa = 7.24NLTNTGNGDD440 pKa = 3.6EE441 pKa = 4.66NLTLSLTQTEE451 pKa = 4.31SDD453 pKa = 3.66FTVTYY458 pKa = 10.23IEE460 pKa = 4.89YY461 pKa = 11.08YY462 pKa = 10.88NDD464 pKa = 3.51ANGNGVEE471 pKa = 5.02DD472 pKa = 4.09LGEE475 pKa = 4.26EE476 pKa = 4.22NLTGGPLALGAGEE489 pKa = 4.84SINLLVKK496 pKa = 10.55IGLPTGLDD504 pKa = 3.54DD505 pKa = 6.02NIATGEE511 pKa = 4.12INATTTSCAVNKK523 pKa = 10.43SAVVYY528 pKa = 9.82DD529 pKa = 4.11QIRR532 pKa = 11.84VLPLNFDD539 pKa = 3.89LDD541 pKa = 3.64ITKK544 pKa = 10.3EE545 pKa = 4.08VSDD548 pKa = 5.72DD549 pKa = 3.45NASWVDD555 pKa = 3.83TIEE558 pKa = 4.29LSEE561 pKa = 4.51GSPAYY566 pKa = 10.7YY567 pKa = 10.44RR568 pKa = 11.84ITVTNNGPDD577 pKa = 3.32GATDD581 pKa = 3.36VTVNDD586 pKa = 3.7VLPANVNYY594 pKa = 9.86IGYY597 pKa = 9.28YY598 pKa = 8.26ATQGTYY604 pKa = 10.78GAGQWMVGNLALDD617 pKa = 4.42GSAEE621 pKa = 4.01LFIDD625 pKa = 3.85VNVSQTAGDD634 pKa = 3.58LTVIQNIASTTDD646 pKa = 3.19TNDD649 pKa = 3.05TDD651 pKa = 5.16TNDD654 pKa = 3.43QDD656 pKa = 4.34DD657 pKa = 4.36ANITAIHH664 pKa = 6.26IPVVNLQIVKK674 pKa = 8.8TVSNPTPYY682 pKa = 10.31EE683 pKa = 4.25GEE685 pKa = 4.51DD686 pKa = 3.05INYY689 pKa = 7.68TITVTKK695 pKa = 10.49FGIDD699 pKa = 3.31DD700 pKa = 4.01AEE702 pKa = 3.99NVVVNDD708 pKa = 4.34ILPPWVSYY716 pKa = 11.18VDD718 pKa = 3.59STASVGSYY726 pKa = 11.14DD727 pKa = 3.92EE728 pKa = 4.44TTGIWDD734 pKa = 3.77IGTMSNSVEE743 pKa = 4.08TLIIQATVDD752 pKa = 3.61TNATLHH758 pKa = 5.79NPIVNTASVSTDD770 pKa = 3.17TTDD773 pKa = 3.16TNPDD777 pKa = 3.59DD778 pKa = 6.13DD779 pKa = 4.07IASATFNASNPADD792 pKa = 3.86LGITKK797 pKa = 9.07TVDD800 pKa = 3.12NPNPQEE806 pKa = 4.08GDD808 pKa = 3.67TIVYY812 pKa = 7.58TISVTNYY819 pKa = 9.91GPLDD823 pKa = 3.44ATNIYY828 pKa = 9.02VQEE831 pKa = 4.16EE832 pKa = 4.43PFDD835 pKa = 4.44GNLSITGTDD844 pKa = 3.22TTLGTFNAPWWHH856 pKa = 5.91IPEE859 pKa = 4.96LNVSQTAVLTVTAMVDD875 pKa = 3.63LNSTGDD881 pKa = 3.47ILINNVIIDD890 pKa = 4.09TEE892 pKa = 4.11RR893 pKa = 11.84LDD895 pKa = 4.22QNDD898 pKa = 3.99TNGEE902 pKa = 3.93NDD904 pKa = 3.27KK905 pKa = 10.18ATAIVIVGCPCDD917 pKa = 4.58NISSDD922 pKa = 4.11SASAMNKK929 pKa = 8.49TVGLLMICMTLLIGLIFVRR948 pKa = 11.84RR949 pKa = 11.84EE950 pKa = 3.34EE951 pKa = 4.06KK952 pKa = 10.45YY953 pKa = 10.82NRR955 pKa = 11.84NEE957 pKa = 3.7RR958 pKa = 3.68
Molecular weight: 102.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I2K7J7|I2K7J7_9PROT ATP synthase subunit a OS=Sulfurovum sp. AR OX=1165841 GN=atpB PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 9.96QPHH8 pKa = 4.86STPRR12 pKa = 11.84KK13 pKa = 7.0RR14 pKa = 11.84THH16 pKa = 6.38GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.87TKK25 pKa = 10.11NGRR28 pKa = 11.84KK29 pKa = 9.33VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 9.96QPHH8 pKa = 4.86STPRR12 pKa = 11.84KK13 pKa = 7.0RR14 pKa = 11.84THH16 pKa = 6.38GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.87TKK25 pKa = 10.11NGRR28 pKa = 11.84KK29 pKa = 9.33VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
643284 |
29 |
2708 |
304.3 |
34.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.284 ± 0.065 | 0.872 ± 0.018 |
5.736 ± 0.048 | 7.278 ± 0.06 |
4.686 ± 0.038 | 6.454 ± 0.055 |
2.192 ± 0.027 | 7.859 ± 0.049 |
7.619 ± 0.064 | 9.77 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.89 ± 0.027 | 4.356 ± 0.043 |
3.348 ± 0.031 | 3.093 ± 0.029 |
3.627 ± 0.036 | 6.102 ± 0.036 |
5.528 ± 0.045 | 6.503 ± 0.048 |
0.879 ± 0.02 | 3.925 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
