
Syncephalastrum racemosum (Filamentous fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Syncephalastraceae; Syncephalastrum
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 11037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
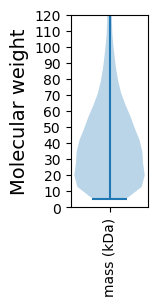
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1X2HQI1|A0A1X2HQI1_SYNRA Helicase C-terminal domain-containing protein (Fragment) OS=Syncephalastrum racemosum OX=13706 GN=BCR43DRAFT_542597 PE=4 SV=1
MM1 pKa = 7.33MPPLAKK7 pKa = 10.11TMVAAAAALLLGGNAVMSAPTNTAFPTGAPSAAASASGASAGASVAAKK55 pKa = 10.05ASKK58 pKa = 10.65GAGNSDD64 pKa = 3.67DD65 pKa = 5.34AFTNSTTPQVQITSDD80 pKa = 3.6TNFCLFMTPQPGLEE94 pKa = 3.97VATNEE99 pKa = 3.96EE100 pKa = 4.14NGIPFCTQANGVPNAKK116 pKa = 8.74TFPEE120 pKa = 4.48GFITTAHH127 pKa = 6.26YY128 pKa = 8.04EE129 pKa = 3.77QSTTYY134 pKa = 10.38VQVTGFFDD142 pKa = 3.34HH143 pKa = 6.78TKK145 pKa = 10.61YY146 pKa = 11.06GYY148 pKa = 11.13ADD150 pKa = 4.27DD151 pKa = 6.01DD152 pKa = 4.38GGGQYY157 pKa = 10.65DD158 pKa = 3.27SHH160 pKa = 8.04GNGKK164 pKa = 9.0PIAASCVGYY173 pKa = 9.33PYY175 pKa = 10.31FVSLLEE181 pKa = 3.89PSNNRR186 pKa = 11.84FCIRR190 pKa = 11.84CCQQEE195 pKa = 4.12DD196 pKa = 3.87DD197 pKa = 4.68CPTGRR202 pKa = 11.84SQYY205 pKa = 10.2GCLRR209 pKa = 11.84VIPGDD214 pKa = 3.68YY215 pKa = 9.51TQGPSSFDD223 pKa = 3.16NDD225 pKa = 3.18QAAGVLNKK233 pKa = 9.91VMEE236 pKa = 4.8DD237 pKa = 3.7FPTYY241 pKa = 10.1GQSIQQQGEE250 pKa = 4.11LDD252 pKa = 3.71EE253 pKa = 5.74LEE255 pKa = 4.19QAVAANTSAVQVQRR269 pKa = 11.84GFSNFNRR276 pKa = 11.84GLADD280 pKa = 3.48QNPNAQQDD288 pKa = 4.04LTQLTDD294 pKa = 3.51FTGNFSSVAQWTQFVQLLKK313 pKa = 10.33DD314 pKa = 3.83TIDD317 pKa = 3.96GGSANNDD324 pKa = 3.57QPSDD328 pKa = 3.69AAPSDD333 pKa = 3.88AAPSDD338 pKa = 3.88AAPSDD343 pKa = 3.88AAPSDD348 pKa = 3.88AAPSDD353 pKa = 3.75AAPSDD358 pKa = 3.97VAPSDD363 pKa = 3.8AAPSDD368 pKa = 3.88AAPSDD373 pKa = 3.89AAPSDD378 pKa = 3.96AQPSDD383 pKa = 3.25AQAPAGQPSDD393 pKa = 4.12AQPSAADD400 pKa = 3.98PSADD404 pKa = 3.61PAGGAPATSSPAAGAQEE421 pKa = 4.57PPAQQQQQQQQQPQQQQQQQPAAPAPAAAPPAAGAAAAPPQQ462 pKa = 3.55
MM1 pKa = 7.33MPPLAKK7 pKa = 10.11TMVAAAAALLLGGNAVMSAPTNTAFPTGAPSAAASASGASAGASVAAKK55 pKa = 10.05ASKK58 pKa = 10.65GAGNSDD64 pKa = 3.67DD65 pKa = 5.34AFTNSTTPQVQITSDD80 pKa = 3.6TNFCLFMTPQPGLEE94 pKa = 3.97VATNEE99 pKa = 3.96EE100 pKa = 4.14NGIPFCTQANGVPNAKK116 pKa = 8.74TFPEE120 pKa = 4.48GFITTAHH127 pKa = 6.26YY128 pKa = 8.04EE129 pKa = 3.77QSTTYY134 pKa = 10.38VQVTGFFDD142 pKa = 3.34HH143 pKa = 6.78TKK145 pKa = 10.61YY146 pKa = 11.06GYY148 pKa = 11.13ADD150 pKa = 4.27DD151 pKa = 6.01DD152 pKa = 4.38GGGQYY157 pKa = 10.65DD158 pKa = 3.27SHH160 pKa = 8.04GNGKK164 pKa = 9.0PIAASCVGYY173 pKa = 9.33PYY175 pKa = 10.31FVSLLEE181 pKa = 3.89PSNNRR186 pKa = 11.84FCIRR190 pKa = 11.84CCQQEE195 pKa = 4.12DD196 pKa = 3.87DD197 pKa = 4.68CPTGRR202 pKa = 11.84SQYY205 pKa = 10.2GCLRR209 pKa = 11.84VIPGDD214 pKa = 3.68YY215 pKa = 9.51TQGPSSFDD223 pKa = 3.16NDD225 pKa = 3.18QAAGVLNKK233 pKa = 9.91VMEE236 pKa = 4.8DD237 pKa = 3.7FPTYY241 pKa = 10.1GQSIQQQGEE250 pKa = 4.11LDD252 pKa = 3.71EE253 pKa = 5.74LEE255 pKa = 4.19QAVAANTSAVQVQRR269 pKa = 11.84GFSNFNRR276 pKa = 11.84GLADD280 pKa = 3.48QNPNAQQDD288 pKa = 4.04LTQLTDD294 pKa = 3.51FTGNFSSVAQWTQFVQLLKK313 pKa = 10.33DD314 pKa = 3.83TIDD317 pKa = 3.96GGSANNDD324 pKa = 3.57QPSDD328 pKa = 3.69AAPSDD333 pKa = 3.88AAPSDD338 pKa = 3.88AAPSDD343 pKa = 3.88AAPSDD348 pKa = 3.88AAPSDD353 pKa = 3.75AAPSDD358 pKa = 3.97VAPSDD363 pKa = 3.8AAPSDD368 pKa = 3.88AAPSDD373 pKa = 3.89AAPSDD378 pKa = 3.96AQPSDD383 pKa = 3.25AQAPAGQPSDD393 pKa = 4.12AQPSAADD400 pKa = 3.98PSADD404 pKa = 3.61PAGGAPATSSPAAGAQEE421 pKa = 4.57PPAQQQQQQQQQPQQQQQQQPAAPAPAAAPPAAGAAAAPPQQ462 pKa = 3.55
Molecular weight: 46.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X2HLV1|A0A1X2HLV1_SYNRA TNase-like domain-containing protein OS=Syncephalastrum racemosum OX=13706 GN=BCR43DRAFT_434868 PE=4 SV=1
MM1 pKa = 7.19VLRR4 pKa = 11.84RR5 pKa = 11.84SPRR8 pKa = 11.84RR9 pKa = 11.84LPLRR13 pKa = 11.84LPPRR17 pKa = 11.84RR18 pKa = 11.84LLRR21 pKa = 11.84HH22 pKa = 6.22RR23 pKa = 11.84LLLHH27 pKa = 6.29HH28 pKa = 7.08RR29 pKa = 11.84RR30 pKa = 11.84LLPQWLPRR38 pKa = 11.84QLLPPQLLLLLLLVHH53 pKa = 7.02RR54 pKa = 11.84LPLPRR59 pKa = 11.84LPVRR63 pKa = 11.84RR64 pKa = 11.84LLRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84LPLRR73 pKa = 11.84PPSLNLPATSARR85 pKa = 11.84LRR87 pKa = 11.84LHH89 pKa = 6.85RR90 pKa = 11.84LLLLLHH96 pKa = 6.8RR97 pKa = 11.84LLLRR101 pKa = 11.84LRR103 pKa = 11.84PILSLPLRR111 pKa = 11.84RR112 pKa = 11.84PSTHH116 pKa = 5.9PRR118 pKa = 11.84HH119 pKa = 6.54RR120 pKa = 11.84ARR122 pKa = 11.84QLILRR127 pKa = 11.84NVAKK131 pKa = 10.14RR132 pKa = 11.84ASSSSSLRR140 pKa = 11.84CQRR143 pKa = 11.84MLWRR147 pKa = 11.84AA148 pKa = 3.14
MM1 pKa = 7.19VLRR4 pKa = 11.84RR5 pKa = 11.84SPRR8 pKa = 11.84RR9 pKa = 11.84LPLRR13 pKa = 11.84LPPRR17 pKa = 11.84RR18 pKa = 11.84LLRR21 pKa = 11.84HH22 pKa = 6.22RR23 pKa = 11.84LLLHH27 pKa = 6.29HH28 pKa = 7.08RR29 pKa = 11.84RR30 pKa = 11.84LLPQWLPRR38 pKa = 11.84QLLPPQLLLLLLLVHH53 pKa = 7.02RR54 pKa = 11.84LPLPRR59 pKa = 11.84LPVRR63 pKa = 11.84RR64 pKa = 11.84LLRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84LPLRR73 pKa = 11.84PPSLNLPATSARR85 pKa = 11.84LRR87 pKa = 11.84LHH89 pKa = 6.85RR90 pKa = 11.84LLLLLHH96 pKa = 6.8RR97 pKa = 11.84LLLRR101 pKa = 11.84LRR103 pKa = 11.84PILSLPLRR111 pKa = 11.84RR112 pKa = 11.84PSTHH116 pKa = 5.9PRR118 pKa = 11.84HH119 pKa = 6.54RR120 pKa = 11.84ARR122 pKa = 11.84QLILRR127 pKa = 11.84NVAKK131 pKa = 10.14RR132 pKa = 11.84ASSSSSLRR140 pKa = 11.84CQRR143 pKa = 11.84MLWRR147 pKa = 11.84AA148 pKa = 3.14
Molecular weight: 17.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4486539 |
49 |
6660 |
406.5 |
45.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.173 ± 0.025 | 1.436 ± 0.009 |
5.886 ± 0.016 | 6.118 ± 0.024 |
3.773 ± 0.016 | 5.542 ± 0.021 |
2.838 ± 0.012 | 4.926 ± 0.017 |
5.181 ± 0.023 | 9.142 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.516 ± 0.01 | 3.787 ± 0.014 |
5.578 ± 0.027 | 4.863 ± 0.024 |
5.991 ± 0.018 | 7.823 ± 0.029 |
5.954 ± 0.016 | 6.115 ± 0.018 |
1.234 ± 0.008 | 3.126 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
