
Limihaloglobus sulfuriphilus
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Phycisphaerae; Sedimentisphaerales; Sedimentisphaeraceae; Limihaloglobus
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
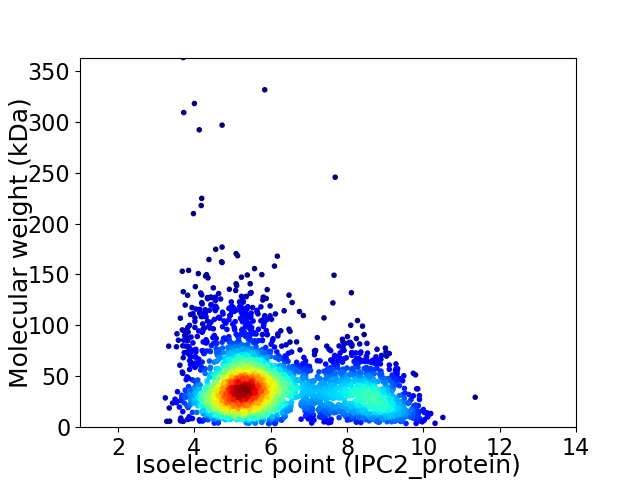
Virtual 2D-PAGE plot for 2870 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q2ME07|A0A1Q2ME07_9BACT Endo-beta-mannanase OS=Limihaloglobus sulfuriphilus OX=1851148 GN=SMSP2_01289 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.38LFQILTMFFFSLTLAVAVSADD23 pKa = 3.67LPAVNSGDD31 pKa = 3.21IFTHH35 pKa = 5.99VSADD39 pKa = 3.22SGVYY43 pKa = 8.79IQQGGSQASDD53 pKa = 3.41GDD55 pKa = 4.33LVGYY59 pKa = 8.62WQDD62 pKa = 3.46QSGNGNNANNFDD74 pKa = 4.02STGEE78 pKa = 4.14SKK80 pKa = 9.12PTFLDD85 pKa = 3.25NAVNGKK91 pKa = 8.84PVLRR95 pKa = 11.84FFGANYY101 pKa = 10.54LDD103 pKa = 4.59FGFNQYY109 pKa = 7.88LTQPNTVFLVAYY121 pKa = 9.97NLGEE125 pKa = 3.81NGYY128 pKa = 9.57YY129 pKa = 9.42IDD131 pKa = 6.52GITSDD136 pKa = 3.99DD137 pKa = 3.43RR138 pKa = 11.84QAFMVADD145 pKa = 4.58GLNVMWSGSYY155 pKa = 9.7FWQDD159 pKa = 2.64HH160 pKa = 5.62SKK162 pKa = 9.55PRR164 pKa = 11.84TWINHH169 pKa = 4.38TLVYY173 pKa = 10.73NDD175 pKa = 4.36TSSQYY180 pKa = 8.71YY181 pKa = 8.82TDD183 pKa = 4.32GVLQAAGSAGASNFGWGLRR202 pKa = 11.84LGSRR206 pKa = 11.84YY207 pKa = 10.15SGSLYY212 pKa = 9.98GWFYY216 pKa = 8.26VAEE219 pKa = 4.25MIIYY223 pKa = 8.98EE224 pKa = 4.3GSLTEE229 pKa = 4.75SDD231 pKa = 2.92ITLVNDD237 pKa = 3.77YY238 pKa = 10.92LSQKK242 pKa = 10.52YY243 pKa = 10.4NITGEE248 pKa = 3.89EE249 pKa = 4.15SQPVNTGAGTLVAQFDD265 pKa = 4.35GTDD268 pKa = 3.07ITADD272 pKa = 3.54GTNVLVWNDD281 pKa = 3.26QVDD284 pKa = 4.02DD285 pKa = 3.56VDD287 pKa = 4.29YY288 pKa = 10.65PLDD291 pKa = 3.77LAAVEE296 pKa = 4.38VDD298 pKa = 3.27MGDD301 pKa = 3.06ASGYY305 pKa = 8.56PQLEE309 pKa = 3.92QYY311 pKa = 11.38DD312 pKa = 3.96FGEE315 pKa = 4.51GSGLKK320 pKa = 10.18NVIRR324 pKa = 11.84FNLPEE329 pKa = 4.57GSSSGTHH336 pKa = 5.63LQSSAIPSTQAGPTTFFVVARR357 pKa = 11.84RR358 pKa = 11.84DD359 pKa = 3.43THH361 pKa = 6.4GFSSYY366 pKa = 11.01LFDD369 pKa = 5.93GISDD373 pKa = 3.59NARR376 pKa = 11.84RR377 pKa = 11.84GVSANGLTDD386 pKa = 2.98THH388 pKa = 6.99RR389 pKa = 11.84LQVPEE394 pKa = 4.13FDD396 pKa = 3.74SVGVDD401 pKa = 3.02VGVWQVFSAVFNEE414 pKa = 4.24EE415 pKa = 3.77FSQLFIDD422 pKa = 5.69GFTADD427 pKa = 3.83EE428 pKa = 4.43YY429 pKa = 11.62GFSGVDD435 pKa = 3.01AFEE438 pKa = 4.44GLTVGSRR445 pKa = 11.84YY446 pKa = 10.15SGSNTMDD453 pKa = 2.6GWIAEE458 pKa = 4.14LLVYY462 pKa = 10.47EE463 pKa = 4.89GALDD467 pKa = 3.93ADD469 pKa = 3.64SRR471 pKa = 11.84KK472 pKa = 9.99AVEE475 pKa = 3.7ASLMQKK481 pKa = 10.63YY482 pKa = 10.45NITPFCGASYY492 pKa = 8.78TQYY495 pKa = 11.56SPMDD499 pKa = 3.65FDD501 pKa = 5.21KK502 pKa = 11.52NCIVNFMDD510 pKa = 3.75YY511 pKa = 11.41AKK513 pKa = 10.53FGQSWLDD520 pKa = 3.46DD521 pKa = 3.77FASIGISSLSGIKK534 pKa = 9.95PSADD538 pKa = 3.3FAHH541 pKa = 6.49GWEE544 pKa = 4.01MDD546 pKa = 3.68YY547 pKa = 11.37YY548 pKa = 9.81PWDD551 pKa = 3.49VANGVDD557 pKa = 4.46EE558 pKa = 6.19DD559 pKa = 4.57NDD561 pKa = 4.12GQIDD565 pKa = 3.89WVRR568 pKa = 11.84SSSNHH573 pKa = 5.82FEE575 pKa = 4.17LSTEE579 pKa = 4.07GTLVNISDD587 pKa = 3.82VSGSLLANDD596 pKa = 5.31DD597 pKa = 3.74NWDD600 pKa = 3.53QAFWPQPDD608 pKa = 3.2INAATGFTTEE618 pKa = 4.24FSIKK622 pKa = 8.76ITSDD626 pKa = 2.86TGSNGAFKK634 pKa = 10.45IQVSPYY640 pKa = 10.45DD641 pKa = 3.5SVNTDD646 pKa = 3.03VVFIGASHH654 pKa = 4.98VTYY657 pKa = 10.84YY658 pKa = 11.18VDD660 pKa = 3.68GGDD663 pKa = 4.45NIVLDD668 pKa = 4.26TSDD671 pKa = 3.52NTDD674 pKa = 3.03GFHH677 pKa = 6.75SFRR680 pKa = 11.84IARR683 pKa = 11.84HH684 pKa = 4.61VDD686 pKa = 3.25EE687 pKa = 4.7PTKK690 pKa = 10.94SSVWKK695 pKa = 10.59DD696 pKa = 3.23GVLIGDD702 pKa = 4.31SLDD705 pKa = 3.88VTWTPSGPHH714 pKa = 6.68RR715 pKa = 11.84YY716 pKa = 9.75YY717 pKa = 10.33IGDD720 pKa = 3.2IDD722 pKa = 4.49SSVNGTYY729 pKa = 10.87EE730 pKa = 3.81MDD732 pKa = 3.95YY733 pKa = 11.37LRR735 pKa = 11.84IDD737 pKa = 3.68PGAWAAPAPMDD748 pKa = 4.02FDD750 pKa = 4.1EE751 pKa = 6.2DD752 pKa = 4.94GIVGSKK758 pKa = 10.02DD759 pKa = 3.37LKK761 pKa = 11.17SFALDD766 pKa = 3.37WLNDD770 pKa = 3.54TDD772 pKa = 4.29PANIEE777 pKa = 3.95
MM1 pKa = 7.74KK2 pKa = 10.38LFQILTMFFFSLTLAVAVSADD23 pKa = 3.67LPAVNSGDD31 pKa = 3.21IFTHH35 pKa = 5.99VSADD39 pKa = 3.22SGVYY43 pKa = 8.79IQQGGSQASDD53 pKa = 3.41GDD55 pKa = 4.33LVGYY59 pKa = 8.62WQDD62 pKa = 3.46QSGNGNNANNFDD74 pKa = 4.02STGEE78 pKa = 4.14SKK80 pKa = 9.12PTFLDD85 pKa = 3.25NAVNGKK91 pKa = 8.84PVLRR95 pKa = 11.84FFGANYY101 pKa = 10.54LDD103 pKa = 4.59FGFNQYY109 pKa = 7.88LTQPNTVFLVAYY121 pKa = 9.97NLGEE125 pKa = 3.81NGYY128 pKa = 9.57YY129 pKa = 9.42IDD131 pKa = 6.52GITSDD136 pKa = 3.99DD137 pKa = 3.43RR138 pKa = 11.84QAFMVADD145 pKa = 4.58GLNVMWSGSYY155 pKa = 9.7FWQDD159 pKa = 2.64HH160 pKa = 5.62SKK162 pKa = 9.55PRR164 pKa = 11.84TWINHH169 pKa = 4.38TLVYY173 pKa = 10.73NDD175 pKa = 4.36TSSQYY180 pKa = 8.71YY181 pKa = 8.82TDD183 pKa = 4.32GVLQAAGSAGASNFGWGLRR202 pKa = 11.84LGSRR206 pKa = 11.84YY207 pKa = 10.15SGSLYY212 pKa = 9.98GWFYY216 pKa = 8.26VAEE219 pKa = 4.25MIIYY223 pKa = 8.98EE224 pKa = 4.3GSLTEE229 pKa = 4.75SDD231 pKa = 2.92ITLVNDD237 pKa = 3.77YY238 pKa = 10.92LSQKK242 pKa = 10.52YY243 pKa = 10.4NITGEE248 pKa = 3.89EE249 pKa = 4.15SQPVNTGAGTLVAQFDD265 pKa = 4.35GTDD268 pKa = 3.07ITADD272 pKa = 3.54GTNVLVWNDD281 pKa = 3.26QVDD284 pKa = 4.02DD285 pKa = 3.56VDD287 pKa = 4.29YY288 pKa = 10.65PLDD291 pKa = 3.77LAAVEE296 pKa = 4.38VDD298 pKa = 3.27MGDD301 pKa = 3.06ASGYY305 pKa = 8.56PQLEE309 pKa = 3.92QYY311 pKa = 11.38DD312 pKa = 3.96FGEE315 pKa = 4.51GSGLKK320 pKa = 10.18NVIRR324 pKa = 11.84FNLPEE329 pKa = 4.57GSSSGTHH336 pKa = 5.63LQSSAIPSTQAGPTTFFVVARR357 pKa = 11.84RR358 pKa = 11.84DD359 pKa = 3.43THH361 pKa = 6.4GFSSYY366 pKa = 11.01LFDD369 pKa = 5.93GISDD373 pKa = 3.59NARR376 pKa = 11.84RR377 pKa = 11.84GVSANGLTDD386 pKa = 2.98THH388 pKa = 6.99RR389 pKa = 11.84LQVPEE394 pKa = 4.13FDD396 pKa = 3.74SVGVDD401 pKa = 3.02VGVWQVFSAVFNEE414 pKa = 4.24EE415 pKa = 3.77FSQLFIDD422 pKa = 5.69GFTADD427 pKa = 3.83EE428 pKa = 4.43YY429 pKa = 11.62GFSGVDD435 pKa = 3.01AFEE438 pKa = 4.44GLTVGSRR445 pKa = 11.84YY446 pKa = 10.15SGSNTMDD453 pKa = 2.6GWIAEE458 pKa = 4.14LLVYY462 pKa = 10.47EE463 pKa = 4.89GALDD467 pKa = 3.93ADD469 pKa = 3.64SRR471 pKa = 11.84KK472 pKa = 9.99AVEE475 pKa = 3.7ASLMQKK481 pKa = 10.63YY482 pKa = 10.45NITPFCGASYY492 pKa = 8.78TQYY495 pKa = 11.56SPMDD499 pKa = 3.65FDD501 pKa = 5.21KK502 pKa = 11.52NCIVNFMDD510 pKa = 3.75YY511 pKa = 11.41AKK513 pKa = 10.53FGQSWLDD520 pKa = 3.46DD521 pKa = 3.77FASIGISSLSGIKK534 pKa = 9.95PSADD538 pKa = 3.3FAHH541 pKa = 6.49GWEE544 pKa = 4.01MDD546 pKa = 3.68YY547 pKa = 11.37YY548 pKa = 9.81PWDD551 pKa = 3.49VANGVDD557 pKa = 4.46EE558 pKa = 6.19DD559 pKa = 4.57NDD561 pKa = 4.12GQIDD565 pKa = 3.89WVRR568 pKa = 11.84SSSNHH573 pKa = 5.82FEE575 pKa = 4.17LSTEE579 pKa = 4.07GTLVNISDD587 pKa = 3.82VSGSLLANDD596 pKa = 5.31DD597 pKa = 3.74NWDD600 pKa = 3.53QAFWPQPDD608 pKa = 3.2INAATGFTTEE618 pKa = 4.24FSIKK622 pKa = 8.76ITSDD626 pKa = 2.86TGSNGAFKK634 pKa = 10.45IQVSPYY640 pKa = 10.45DD641 pKa = 3.5SVNTDD646 pKa = 3.03VVFIGASHH654 pKa = 4.98VTYY657 pKa = 10.84YY658 pKa = 11.18VDD660 pKa = 3.68GGDD663 pKa = 4.45NIVLDD668 pKa = 4.26TSDD671 pKa = 3.52NTDD674 pKa = 3.03GFHH677 pKa = 6.75SFRR680 pKa = 11.84IARR683 pKa = 11.84HH684 pKa = 4.61VDD686 pKa = 3.25EE687 pKa = 4.7PTKK690 pKa = 10.94SSVWKK695 pKa = 10.59DD696 pKa = 3.23GVLIGDD702 pKa = 4.31SLDD705 pKa = 3.88VTWTPSGPHH714 pKa = 6.68RR715 pKa = 11.84YY716 pKa = 9.75YY717 pKa = 10.33IGDD720 pKa = 3.2IDD722 pKa = 4.49SSVNGTYY729 pKa = 10.87EE730 pKa = 3.81MDD732 pKa = 3.95YY733 pKa = 11.37LRR735 pKa = 11.84IDD737 pKa = 3.68PGAWAAPAPMDD748 pKa = 4.02FDD750 pKa = 4.1EE751 pKa = 6.2DD752 pKa = 4.94GIVGSKK758 pKa = 10.02DD759 pKa = 3.37LKK761 pKa = 11.17SFALDD766 pKa = 3.37WLNDD770 pKa = 3.54TDD772 pKa = 4.29PANIEE777 pKa = 3.95
Molecular weight: 84.95 kDa
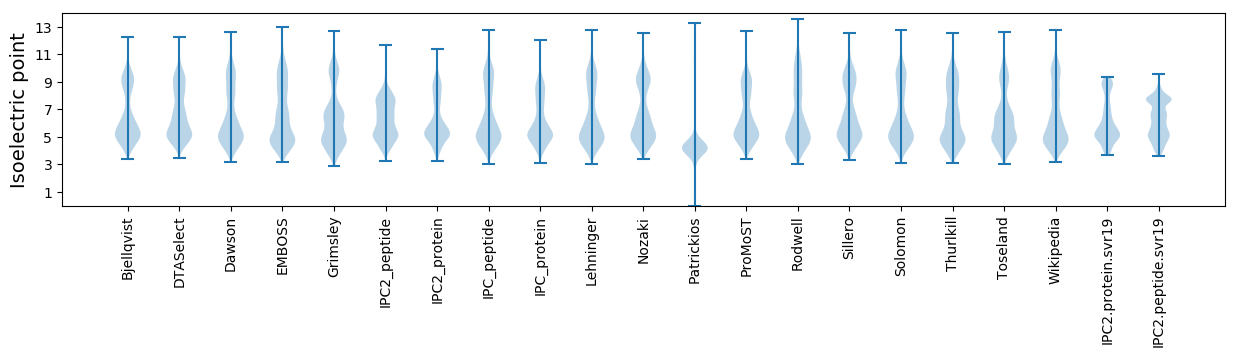
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q2MFI5|A0A1Q2MFI5_9BACT Ribulokinase OS=Limihaloglobus sulfuriphilus OX=1851148 GN=araB PE=3 SV=1
MM1 pKa = 7.56EE2 pKa = 5.76FDD4 pKa = 5.77KK5 pKa = 11.27ASQWLLKK12 pKa = 9.79SLRR15 pKa = 11.84RR16 pKa = 11.84MKK18 pKa = 10.49LLKK21 pKa = 10.32QLAQPMTAFQLYY33 pKa = 9.33KK34 pKa = 9.56RR35 pKa = 11.84TGISRR40 pKa = 11.84RR41 pKa = 11.84DD42 pKa = 3.54CSDD45 pKa = 3.76VIVEE49 pKa = 4.26MYY51 pKa = 10.47KK52 pKa = 10.71AGLLFCLNPKK62 pKa = 8.62ATRR65 pKa = 11.84SRR67 pKa = 11.84LFGLTKK73 pKa = 9.53TGKK76 pKa = 10.11SIRR79 pKa = 11.84KK80 pKa = 8.35QLFPRR85 pKa = 11.84IEE87 pKa = 4.29PYY89 pKa = 10.36YY90 pKa = 10.74EE91 pKa = 5.11PDD93 pKa = 3.43IDD95 pKa = 3.62WNAYY99 pKa = 10.04GYY101 pKa = 11.09VCFSQRR107 pKa = 11.84SAIILALDD115 pKa = 3.91SPRR118 pKa = 11.84SPAGIKK124 pKa = 9.95RR125 pKa = 11.84YY126 pKa = 9.31LRR128 pKa = 11.84HH129 pKa = 6.54ILPKK133 pKa = 10.19TRR135 pKa = 11.84ISANNIRR142 pKa = 11.84NNIPMLKK149 pKa = 10.06ARR151 pKa = 11.84DD152 pKa = 3.78IIRR155 pKa = 11.84TIDD158 pKa = 3.23TRR160 pKa = 11.84GVYY163 pKa = 9.67PEE165 pKa = 4.45YY166 pKa = 10.66EE167 pKa = 4.08LTVLGRR173 pKa = 11.84QLQTLLRR180 pKa = 11.84RR181 pKa = 11.84TASPSQQ187 pKa = 3.19
MM1 pKa = 7.56EE2 pKa = 5.76FDD4 pKa = 5.77KK5 pKa = 11.27ASQWLLKK12 pKa = 9.79SLRR15 pKa = 11.84RR16 pKa = 11.84MKK18 pKa = 10.49LLKK21 pKa = 10.32QLAQPMTAFQLYY33 pKa = 9.33KK34 pKa = 9.56RR35 pKa = 11.84TGISRR40 pKa = 11.84RR41 pKa = 11.84DD42 pKa = 3.54CSDD45 pKa = 3.76VIVEE49 pKa = 4.26MYY51 pKa = 10.47KK52 pKa = 10.71AGLLFCLNPKK62 pKa = 8.62ATRR65 pKa = 11.84SRR67 pKa = 11.84LFGLTKK73 pKa = 9.53TGKK76 pKa = 10.11SIRR79 pKa = 11.84KK80 pKa = 8.35QLFPRR85 pKa = 11.84IEE87 pKa = 4.29PYY89 pKa = 10.36YY90 pKa = 10.74EE91 pKa = 5.11PDD93 pKa = 3.43IDD95 pKa = 3.62WNAYY99 pKa = 10.04GYY101 pKa = 11.09VCFSQRR107 pKa = 11.84SAIILALDD115 pKa = 3.91SPRR118 pKa = 11.84SPAGIKK124 pKa = 9.95RR125 pKa = 11.84YY126 pKa = 9.31LRR128 pKa = 11.84HH129 pKa = 6.54ILPKK133 pKa = 10.19TRR135 pKa = 11.84ISANNIRR142 pKa = 11.84NNIPMLKK149 pKa = 10.06ARR151 pKa = 11.84DD152 pKa = 3.78IIRR155 pKa = 11.84TIDD158 pKa = 3.23TRR160 pKa = 11.84GVYY163 pKa = 9.67PEE165 pKa = 4.45YY166 pKa = 10.66EE167 pKa = 4.08LTVLGRR173 pKa = 11.84QLQTLLRR180 pKa = 11.84RR181 pKa = 11.84TASPSQQ187 pKa = 3.19
Molecular weight: 21.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1126502 |
30 |
3405 |
392.5 |
43.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.856 ± 0.044 | 1.437 ± 0.018 |
6.246 ± 0.037 | 6.363 ± 0.037 |
4.317 ± 0.031 | 7.59 ± 0.043 |
1.793 ± 0.016 | 6.878 ± 0.035 |
5.762 ± 0.051 | 8.655 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.554 ± 0.023 | 4.546 ± 0.032 |
4.19 ± 0.026 | 3.252 ± 0.023 |
4.871 ± 0.038 | 6.718 ± 0.035 |
5.24 ± 0.037 | 6.475 ± 0.034 |
1.471 ± 0.019 | 3.785 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
