
Gemycircularvirus HV-GcV2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas5; Human associated gemykibivirus 5
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
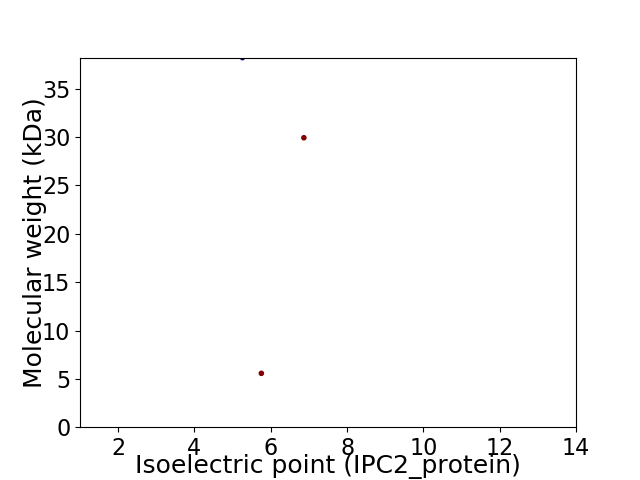
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A193CE78|A0A193CE78_9VIRU Capsid protein OS=Gemycircularvirus HV-GcV2 OX=1862825 GN=cap PE=4 SV=1
MM1 pKa = 7.89LGEE4 pKa = 5.09LGAEE8 pKa = 4.07CIVGRR13 pKa = 11.84EE14 pKa = 3.97SHH16 pKa = 6.96ADD18 pKa = 3.35GGLHH22 pKa = 5.98LHH24 pKa = 6.48AFFMFEE30 pKa = 4.19RR31 pKa = 11.84KK32 pKa = 9.23FEE34 pKa = 4.26SRR36 pKa = 11.84DD37 pKa = 3.27VRR39 pKa = 11.84VFDD42 pKa = 4.91VDD44 pKa = 3.72GLHH47 pKa = 6.49PNIVRR52 pKa = 11.84GYY54 pKa = 4.45TTPRR58 pKa = 11.84KK59 pKa = 9.59GADD62 pKa = 3.33YY63 pKa = 10.49AIKK66 pKa = 10.86DD67 pKa = 3.61GDD69 pKa = 4.09VVAGGLDD76 pKa = 3.47ISQLGAPVSHH86 pKa = 6.97TSAVWATIIMASTRR100 pKa = 11.84EE101 pKa = 4.06EE102 pKa = 4.09FFEE105 pKa = 4.68ACATLAPRR113 pKa = 11.84SLVCSFTSLKK123 pKa = 10.62CYY125 pKa = 10.61ADD127 pKa = 2.9WKK129 pKa = 10.1YY130 pKa = 10.83RR131 pKa = 11.84PEE133 pKa = 4.19PVPYY137 pKa = 9.09EE138 pKa = 4.27HH139 pKa = 7.71PSGLSFDD146 pKa = 3.63TTAYY150 pKa = 9.77PEE152 pKa = 3.99LDD154 pKa = 2.93AWVSHH159 pKa = 5.92ALEE162 pKa = 4.66GPEE165 pKa = 5.0CGGEE169 pKa = 4.1CPPTAEE175 pKa = 4.47DD176 pKa = 3.91TFPSLRR182 pKa = 11.84PPGALRR188 pKa = 11.84VYY190 pKa = 11.12AMSHH194 pKa = 6.03CWAAWRR200 pKa = 11.84LPAKK204 pKa = 10.6LMGVMLTYY212 pKa = 10.36SGRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84SLVLYY222 pKa = 10.58GPTRR226 pKa = 11.84LGKK229 pKa = 7.65TLWARR234 pKa = 11.84ALGSHH239 pKa = 7.14AYY241 pKa = 10.15FGGLFSLDD249 pKa = 3.47EE250 pKa = 4.27SLEE253 pKa = 4.28DD254 pKa = 2.83VDD256 pKa = 5.06YY257 pKa = 11.57AVFDD261 pKa = 4.3DD262 pKa = 4.09MQGGLKK268 pKa = 10.02FFHH271 pKa = 7.28AYY273 pKa = 9.59KK274 pKa = 10.26FWLGAQSQFWATDD287 pKa = 3.02KK288 pKa = 11.5YY289 pKa = 10.22KK290 pKa = 10.83GKK292 pKa = 10.54RR293 pKa = 11.84LIHH296 pKa = 6.0WGKK299 pKa = 9.27PSIYY303 pKa = 9.39IANGNPLTDD312 pKa = 3.55EE313 pKa = 4.88GVDD316 pKa = 3.87HH317 pKa = 7.71DD318 pKa = 4.77WLLGNCDD325 pKa = 3.46FVEE328 pKa = 4.31ITTSLLVPEE337 pKa = 4.86VGPVTLEE344 pKa = 3.85VV345 pKa = 3.38
MM1 pKa = 7.89LGEE4 pKa = 5.09LGAEE8 pKa = 4.07CIVGRR13 pKa = 11.84EE14 pKa = 3.97SHH16 pKa = 6.96ADD18 pKa = 3.35GGLHH22 pKa = 5.98LHH24 pKa = 6.48AFFMFEE30 pKa = 4.19RR31 pKa = 11.84KK32 pKa = 9.23FEE34 pKa = 4.26SRR36 pKa = 11.84DD37 pKa = 3.27VRR39 pKa = 11.84VFDD42 pKa = 4.91VDD44 pKa = 3.72GLHH47 pKa = 6.49PNIVRR52 pKa = 11.84GYY54 pKa = 4.45TTPRR58 pKa = 11.84KK59 pKa = 9.59GADD62 pKa = 3.33YY63 pKa = 10.49AIKK66 pKa = 10.86DD67 pKa = 3.61GDD69 pKa = 4.09VVAGGLDD76 pKa = 3.47ISQLGAPVSHH86 pKa = 6.97TSAVWATIIMASTRR100 pKa = 11.84EE101 pKa = 4.06EE102 pKa = 4.09FFEE105 pKa = 4.68ACATLAPRR113 pKa = 11.84SLVCSFTSLKK123 pKa = 10.62CYY125 pKa = 10.61ADD127 pKa = 2.9WKK129 pKa = 10.1YY130 pKa = 10.83RR131 pKa = 11.84PEE133 pKa = 4.19PVPYY137 pKa = 9.09EE138 pKa = 4.27HH139 pKa = 7.71PSGLSFDD146 pKa = 3.63TTAYY150 pKa = 9.77PEE152 pKa = 3.99LDD154 pKa = 2.93AWVSHH159 pKa = 5.92ALEE162 pKa = 4.66GPEE165 pKa = 5.0CGGEE169 pKa = 4.1CPPTAEE175 pKa = 4.47DD176 pKa = 3.91TFPSLRR182 pKa = 11.84PPGALRR188 pKa = 11.84VYY190 pKa = 11.12AMSHH194 pKa = 6.03CWAAWRR200 pKa = 11.84LPAKK204 pKa = 10.6LMGVMLTYY212 pKa = 10.36SGRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84SLVLYY222 pKa = 10.58GPTRR226 pKa = 11.84LGKK229 pKa = 7.65TLWARR234 pKa = 11.84ALGSHH239 pKa = 7.14AYY241 pKa = 10.15FGGLFSLDD249 pKa = 3.47EE250 pKa = 4.27SLEE253 pKa = 4.28DD254 pKa = 2.83VDD256 pKa = 5.06YY257 pKa = 11.57AVFDD261 pKa = 4.3DD262 pKa = 4.09MQGGLKK268 pKa = 10.02FFHH271 pKa = 7.28AYY273 pKa = 9.59KK274 pKa = 10.26FWLGAQSQFWATDD287 pKa = 3.02KK288 pKa = 11.5YY289 pKa = 10.22KK290 pKa = 10.83GKK292 pKa = 10.54RR293 pKa = 11.84LIHH296 pKa = 6.0WGKK299 pKa = 9.27PSIYY303 pKa = 9.39IANGNPLTDD312 pKa = 3.55EE313 pKa = 4.88GVDD316 pKa = 3.87HH317 pKa = 7.71DD318 pKa = 4.77WLLGNCDD325 pKa = 3.46FVEE328 pKa = 4.31ITTSLLVPEE337 pKa = 4.86VGPVTLEE344 pKa = 3.85VV345 pKa = 3.38
Molecular weight: 38.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A193CE78|A0A193CE78_9VIRU Capsid protein OS=Gemycircularvirus HV-GcV2 OX=1862825 GN=cap PE=4 SV=1
MM1 pKa = 7.15SVRR4 pKa = 11.84RR5 pKa = 11.84ILNKK9 pKa = 9.59TSQKK13 pKa = 10.43KK14 pKa = 9.89RR15 pKa = 11.84DD16 pKa = 3.83TLLTWSNVDD25 pKa = 3.86PSNPDD30 pKa = 3.55PDD32 pKa = 4.01NDD34 pKa = 4.01TYY36 pKa = 10.9TPSGATIPGGTGAFQYY52 pKa = 11.0MMFCPTARR60 pKa = 11.84GLEE63 pKa = 4.29SFTGSLGSKK72 pKa = 10.37ANTTIRR78 pKa = 11.84TSTTVFARR86 pKa = 11.84GYY88 pKa = 10.93KK89 pKa = 10.16EE90 pKa = 3.47ITQLRR95 pKa = 11.84TRR97 pKa = 11.84SGKK100 pKa = 7.9GWQWRR105 pKa = 11.84RR106 pKa = 11.84ICFTLKK112 pKa = 10.52GDD114 pKa = 4.25SLNEE118 pKa = 4.12GNSDD122 pKa = 3.78PASSPVFRR130 pKa = 11.84EE131 pKa = 3.74TSNGYY136 pKa = 9.02QRR138 pKa = 11.84LIKK141 pKa = 10.27IADD144 pKa = 3.96PDD146 pKa = 3.77RR147 pKa = 11.84VSTLLFRR154 pKa = 11.84GNNGQDD160 pKa = 3.06WNDD163 pKa = 3.28PTMASLDD170 pKa = 3.65TRR172 pKa = 11.84RR173 pKa = 11.84VSVKK177 pKa = 9.86YY178 pKa = 10.39DD179 pKa = 3.5RR180 pKa = 11.84LRR182 pKa = 11.84MIRR185 pKa = 11.84SGNDD189 pKa = 3.02DD190 pKa = 3.55GVVRR194 pKa = 11.84TYY196 pKa = 10.81KK197 pKa = 10.06QWFPMNKK204 pKa = 9.43NIYY207 pKa = 10.01YY208 pKa = 10.17EE209 pKa = 4.46DD210 pKa = 4.9DD211 pKa = 3.73EE212 pKa = 5.52SGSTMNTNPFSVEE225 pKa = 4.18SNQGMGDD232 pKa = 3.75YY233 pKa = 10.68YY234 pKa = 10.44IVDD237 pKa = 3.95IFRR240 pKa = 11.84ANPDD244 pKa = 3.13TTAEE248 pKa = 4.12DD249 pKa = 3.77SLDD252 pKa = 3.8FEE254 pKa = 5.95CNGTYY259 pKa = 9.97FWHH262 pKa = 6.74EE263 pKa = 4.0KK264 pKa = 10.03
MM1 pKa = 7.15SVRR4 pKa = 11.84RR5 pKa = 11.84ILNKK9 pKa = 9.59TSQKK13 pKa = 10.43KK14 pKa = 9.89RR15 pKa = 11.84DD16 pKa = 3.83TLLTWSNVDD25 pKa = 3.86PSNPDD30 pKa = 3.55PDD32 pKa = 4.01NDD34 pKa = 4.01TYY36 pKa = 10.9TPSGATIPGGTGAFQYY52 pKa = 11.0MMFCPTARR60 pKa = 11.84GLEE63 pKa = 4.29SFTGSLGSKK72 pKa = 10.37ANTTIRR78 pKa = 11.84TSTTVFARR86 pKa = 11.84GYY88 pKa = 10.93KK89 pKa = 10.16EE90 pKa = 3.47ITQLRR95 pKa = 11.84TRR97 pKa = 11.84SGKK100 pKa = 7.9GWQWRR105 pKa = 11.84RR106 pKa = 11.84ICFTLKK112 pKa = 10.52GDD114 pKa = 4.25SLNEE118 pKa = 4.12GNSDD122 pKa = 3.78PASSPVFRR130 pKa = 11.84EE131 pKa = 3.74TSNGYY136 pKa = 9.02QRR138 pKa = 11.84LIKK141 pKa = 10.27IADD144 pKa = 3.96PDD146 pKa = 3.77RR147 pKa = 11.84VSTLLFRR154 pKa = 11.84GNNGQDD160 pKa = 3.06WNDD163 pKa = 3.28PTMASLDD170 pKa = 3.65TRR172 pKa = 11.84RR173 pKa = 11.84VSVKK177 pKa = 9.86YY178 pKa = 10.39DD179 pKa = 3.5RR180 pKa = 11.84LRR182 pKa = 11.84MIRR185 pKa = 11.84SGNDD189 pKa = 3.02DD190 pKa = 3.55GVVRR194 pKa = 11.84TYY196 pKa = 10.81KK197 pKa = 10.06QWFPMNKK204 pKa = 9.43NIYY207 pKa = 10.01YY208 pKa = 10.17EE209 pKa = 4.46DD210 pKa = 4.9DD211 pKa = 3.73EE212 pKa = 5.52SGSTMNTNPFSVEE225 pKa = 4.18SNQGMGDD232 pKa = 3.75YY233 pKa = 10.68YY234 pKa = 10.44IVDD237 pKa = 3.95IFRR240 pKa = 11.84ANPDD244 pKa = 3.13TTAEE248 pKa = 4.12DD249 pKa = 3.77SLDD252 pKa = 3.8FEE254 pKa = 5.95CNGTYY259 pKa = 9.97FWHH262 pKa = 6.74EE263 pKa = 4.0KK264 pKa = 10.03
Molecular weight: 29.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
663 |
54 |
345 |
221.0 |
24.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.486 ± 1.808 | 1.659 ± 0.533 |
7.391 ± 0.766 | 4.827 ± 1.001 |
4.676 ± 0.625 | 9.653 ± 1.742 |
2.262 ± 0.976 | 3.469 ± 0.41 |
4.072 ± 0.245 | 7.994 ± 1.55 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.715 ± 0.684 | 3.469 ± 1.922 |
5.279 ± 0.817 | 1.961 ± 0.588 |
6.335 ± 0.854 | 7.089 ± 1.235 |
8.446 ± 2.66 | 6.033 ± 1.174 |
2.413 ± 0.542 | 3.771 ± 0.738 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
