
Bromus-associated circular DNA virus 4
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
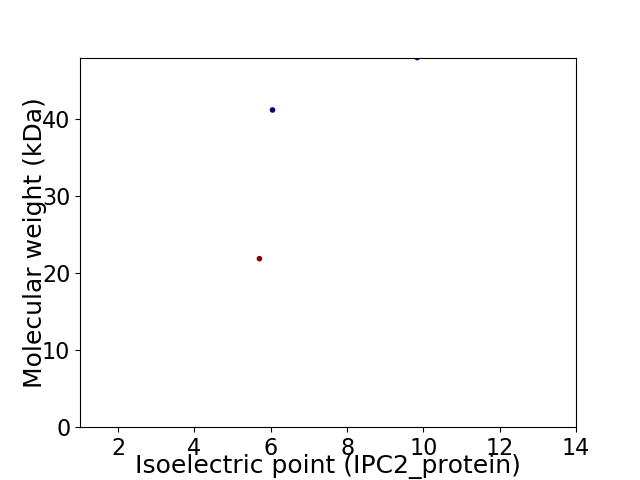
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
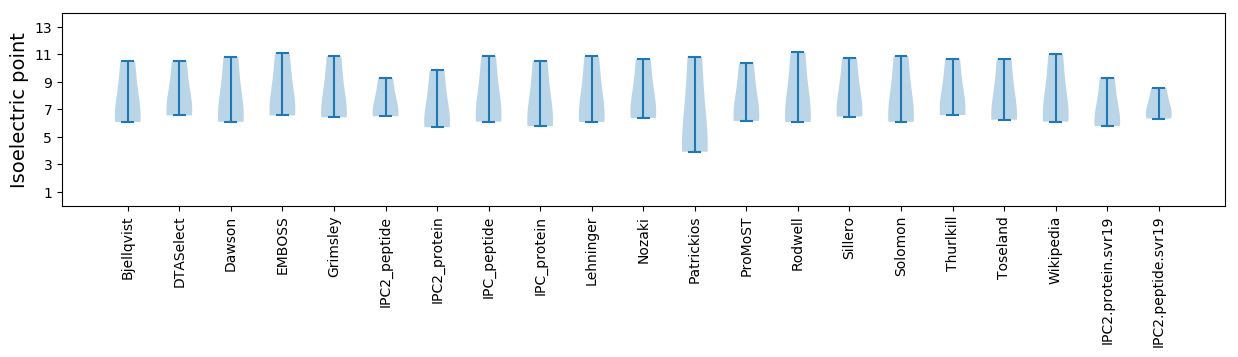
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4U841|A0A0B4U841_9VIRU Replication-associated protein OS=Bromus-associated circular DNA virus 4 OX=1590172 PE=3 SV=1
MM1 pKa = 7.71AGEE4 pKa = 4.12LSEE7 pKa = 4.67FCPHH11 pKa = 6.12SNPWRR16 pKa = 11.84HH17 pKa = 5.54EE18 pKa = 4.17PGEE21 pKa = 4.29GRR23 pKa = 11.84HH24 pKa = 5.31EE25 pKa = 4.04EE26 pKa = 3.95HH27 pKa = 7.24AKK29 pKa = 10.3VCGEE33 pKa = 3.87QDD35 pKa = 3.33SLRR38 pKa = 11.84PADD41 pKa = 4.36PEE43 pKa = 3.97HH44 pKa = 5.87VQPGRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84DD52 pKa = 3.89LIRR55 pKa = 11.84CGYY58 pKa = 8.81EE59 pKa = 3.74EE60 pKa = 4.15QRR62 pKa = 11.84ATQPYY67 pKa = 8.37PRR69 pKa = 11.84NGNEE73 pKa = 3.87LARR76 pKa = 11.84SRR78 pKa = 11.84SVEE81 pKa = 3.7GLGIRR86 pKa = 11.84TSDD89 pKa = 3.47RR90 pKa = 11.84ADD92 pKa = 3.0SGGPLLGDD100 pKa = 4.75PIRR103 pKa = 11.84CHH105 pKa = 6.59WYY107 pKa = 9.57HH108 pKa = 6.45AVHH111 pKa = 6.48VDD113 pKa = 3.54GVLQTVPHH121 pKa = 6.44CKK123 pKa = 9.82GHH125 pKa = 6.01EE126 pKa = 4.04EE127 pKa = 5.16NLVFWCGPSPPHH139 pKa = 6.73YY140 pKa = 10.59SEE142 pKa = 4.03ATADD146 pKa = 3.59VRR148 pKa = 11.84HH149 pKa = 6.32SGEE152 pKa = 4.17AVCGRR157 pKa = 11.84DD158 pKa = 3.52SAGPGHH164 pKa = 6.19NRR166 pKa = 11.84PDD168 pKa = 3.35RR169 pKa = 11.84GFLAWALSPRR179 pKa = 11.84HH180 pKa = 5.53EE181 pKa = 4.79WIHH184 pKa = 6.28DD185 pKa = 3.82LSSVGLYY192 pKa = 7.81WQQQ195 pKa = 2.8
MM1 pKa = 7.71AGEE4 pKa = 4.12LSEE7 pKa = 4.67FCPHH11 pKa = 6.12SNPWRR16 pKa = 11.84HH17 pKa = 5.54EE18 pKa = 4.17PGEE21 pKa = 4.29GRR23 pKa = 11.84HH24 pKa = 5.31EE25 pKa = 4.04EE26 pKa = 3.95HH27 pKa = 7.24AKK29 pKa = 10.3VCGEE33 pKa = 3.87QDD35 pKa = 3.33SLRR38 pKa = 11.84PADD41 pKa = 4.36PEE43 pKa = 3.97HH44 pKa = 5.87VQPGRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84DD52 pKa = 3.89LIRR55 pKa = 11.84CGYY58 pKa = 8.81EE59 pKa = 3.74EE60 pKa = 4.15QRR62 pKa = 11.84ATQPYY67 pKa = 8.37PRR69 pKa = 11.84NGNEE73 pKa = 3.87LARR76 pKa = 11.84SRR78 pKa = 11.84SVEE81 pKa = 3.7GLGIRR86 pKa = 11.84TSDD89 pKa = 3.47RR90 pKa = 11.84ADD92 pKa = 3.0SGGPLLGDD100 pKa = 4.75PIRR103 pKa = 11.84CHH105 pKa = 6.59WYY107 pKa = 9.57HH108 pKa = 6.45AVHH111 pKa = 6.48VDD113 pKa = 3.54GVLQTVPHH121 pKa = 6.44CKK123 pKa = 9.82GHH125 pKa = 6.01EE126 pKa = 4.04EE127 pKa = 5.16NLVFWCGPSPPHH139 pKa = 6.73YY140 pKa = 10.59SEE142 pKa = 4.03ATADD146 pKa = 3.59VRR148 pKa = 11.84HH149 pKa = 6.32SGEE152 pKa = 4.17AVCGRR157 pKa = 11.84DD158 pKa = 3.52SAGPGHH164 pKa = 6.19NRR166 pKa = 11.84PDD168 pKa = 3.35RR169 pKa = 11.84GFLAWALSPRR179 pKa = 11.84HH180 pKa = 5.53EE181 pKa = 4.79WIHH184 pKa = 6.28DD185 pKa = 3.82LSSVGLYY192 pKa = 7.81WQQQ195 pKa = 2.8
Molecular weight: 21.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4U9L1|A0A0B4U9L1_9VIRU Putative capsid protein OS=Bromus-associated circular DNA virus 4 OX=1590172 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84PSHH5 pKa = 5.97GRR7 pKa = 11.84AIPQRR12 pKa = 11.84LHH14 pKa = 6.58AATKK18 pKa = 9.93FAYY21 pKa = 9.6HH22 pKa = 6.88HH23 pKa = 6.55ARR25 pKa = 11.84GAAVKK30 pKa = 10.61AMAKK34 pKa = 9.06YY35 pKa = 9.95ARR37 pKa = 11.84SRR39 pKa = 11.84ATRR42 pKa = 11.84KK43 pKa = 10.01HH44 pKa = 5.42GGGFKK49 pKa = 10.25KK50 pKa = 10.23RR51 pKa = 11.84YY52 pKa = 6.39TPKK55 pKa = 9.93TVLGKK60 pKa = 10.6RR61 pKa = 11.84KK62 pKa = 9.03IASSAGGVSKK72 pKa = 10.54RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 8.31VYY77 pKa = 9.06SSKK80 pKa = 10.67RR81 pKa = 11.84IKK83 pKa = 10.23PYY85 pKa = 10.12SKK87 pKa = 10.19RR88 pKa = 11.84GGKK91 pKa = 9.25RR92 pKa = 11.84RR93 pKa = 11.84GGSFSNAIAKK103 pKa = 9.72VDD105 pKa = 3.53SSAFSLGPGPRR116 pKa = 11.84LFSGFRR122 pKa = 11.84RR123 pKa = 11.84TQQPTTVNCTISSRR137 pKa = 11.84VSSTPSQQAVQLLPTYY153 pKa = 9.86FFAGDD158 pKa = 4.13GSAGDD163 pKa = 4.38PSRR166 pKa = 11.84QSAFTIDD173 pKa = 3.91SYY175 pKa = 11.56ILNRR179 pKa = 11.84AEE181 pKa = 3.37RR182 pKa = 11.84WLANFQNSVPTQTPGGTNQGRR203 pKa = 11.84DD204 pKa = 3.4GMRR207 pKa = 11.84NTQKK211 pKa = 10.48FVVNKK216 pKa = 9.84IHH218 pKa = 7.04YY219 pKa = 7.39DD220 pKa = 3.4QQIRR224 pKa = 11.84NMSNQDD230 pKa = 2.77VDD232 pKa = 4.23VILYY236 pKa = 10.21DD237 pKa = 3.33VVMRR241 pKa = 11.84NNVQPNLTRR250 pKa = 11.84ATGTNLPVADD260 pKa = 4.75PLKK263 pKa = 10.44DD264 pKa = 3.38WEE266 pKa = 4.57SGLLIEE272 pKa = 5.08QIQGGLYY279 pKa = 10.31LGTRR283 pKa = 11.84LGAIGTTPFMSTAFCKK299 pKa = 10.34LYY301 pKa = 10.7RR302 pKa = 11.84IAKK305 pKa = 5.97VTKK308 pKa = 8.33KK309 pKa = 8.41TLSSGAVHH317 pKa = 6.42HH318 pKa = 6.08HH319 pKa = 7.05HH320 pKa = 6.32ITVKK324 pKa = 9.17PRR326 pKa = 11.84QMFDD330 pKa = 3.39TQVKK334 pKa = 9.83RR335 pKa = 11.84YY336 pKa = 9.66ADD338 pKa = 3.66EE339 pKa = 4.16TAPDD343 pKa = 4.48LDD345 pKa = 3.96TTGLIGVSSRR355 pKa = 11.84GPYY358 pKa = 9.77LPGMSGFTILVASGSIGNNNVEE380 pKa = 4.39KK381 pKa = 10.77AQIGYY386 pKa = 8.14TSTALDD392 pKa = 3.82VVTKK396 pKa = 10.78GSFSFSNFTRR406 pKa = 11.84EE407 pKa = 4.33RR408 pKa = 11.84RR409 pKa = 11.84FHH411 pKa = 6.97LNFDD415 pKa = 3.99GLATNVNQIVTDD427 pKa = 3.8DD428 pKa = 3.72TDD430 pKa = 5.08LIGPVQEE437 pKa = 4.28AA438 pKa = 3.48
MM1 pKa = 7.55RR2 pKa = 11.84PSHH5 pKa = 5.97GRR7 pKa = 11.84AIPQRR12 pKa = 11.84LHH14 pKa = 6.58AATKK18 pKa = 9.93FAYY21 pKa = 9.6HH22 pKa = 6.88HH23 pKa = 6.55ARR25 pKa = 11.84GAAVKK30 pKa = 10.61AMAKK34 pKa = 9.06YY35 pKa = 9.95ARR37 pKa = 11.84SRR39 pKa = 11.84ATRR42 pKa = 11.84KK43 pKa = 10.01HH44 pKa = 5.42GGGFKK49 pKa = 10.25KK50 pKa = 10.23RR51 pKa = 11.84YY52 pKa = 6.39TPKK55 pKa = 9.93TVLGKK60 pKa = 10.6RR61 pKa = 11.84KK62 pKa = 9.03IASSAGGVSKK72 pKa = 10.54RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 8.31VYY77 pKa = 9.06SSKK80 pKa = 10.67RR81 pKa = 11.84IKK83 pKa = 10.23PYY85 pKa = 10.12SKK87 pKa = 10.19RR88 pKa = 11.84GGKK91 pKa = 9.25RR92 pKa = 11.84RR93 pKa = 11.84GGSFSNAIAKK103 pKa = 9.72VDD105 pKa = 3.53SSAFSLGPGPRR116 pKa = 11.84LFSGFRR122 pKa = 11.84RR123 pKa = 11.84TQQPTTVNCTISSRR137 pKa = 11.84VSSTPSQQAVQLLPTYY153 pKa = 9.86FFAGDD158 pKa = 4.13GSAGDD163 pKa = 4.38PSRR166 pKa = 11.84QSAFTIDD173 pKa = 3.91SYY175 pKa = 11.56ILNRR179 pKa = 11.84AEE181 pKa = 3.37RR182 pKa = 11.84WLANFQNSVPTQTPGGTNQGRR203 pKa = 11.84DD204 pKa = 3.4GMRR207 pKa = 11.84NTQKK211 pKa = 10.48FVVNKK216 pKa = 9.84IHH218 pKa = 7.04YY219 pKa = 7.39DD220 pKa = 3.4QQIRR224 pKa = 11.84NMSNQDD230 pKa = 2.77VDD232 pKa = 4.23VILYY236 pKa = 10.21DD237 pKa = 3.33VVMRR241 pKa = 11.84NNVQPNLTRR250 pKa = 11.84ATGTNLPVADD260 pKa = 4.75PLKK263 pKa = 10.44DD264 pKa = 3.38WEE266 pKa = 4.57SGLLIEE272 pKa = 5.08QIQGGLYY279 pKa = 10.31LGTRR283 pKa = 11.84LGAIGTTPFMSTAFCKK299 pKa = 10.34LYY301 pKa = 10.7RR302 pKa = 11.84IAKK305 pKa = 5.97VTKK308 pKa = 8.33KK309 pKa = 8.41TLSSGAVHH317 pKa = 6.42HH318 pKa = 6.08HH319 pKa = 7.05HH320 pKa = 6.32ITVKK324 pKa = 9.17PRR326 pKa = 11.84QMFDD330 pKa = 3.39TQVKK334 pKa = 9.83RR335 pKa = 11.84YY336 pKa = 9.66ADD338 pKa = 3.66EE339 pKa = 4.16TAPDD343 pKa = 4.48LDD345 pKa = 3.96TTGLIGVSSRR355 pKa = 11.84GPYY358 pKa = 9.77LPGMSGFTILVASGSIGNNNVEE380 pKa = 4.39KK381 pKa = 10.77AQIGYY386 pKa = 8.14TSTALDD392 pKa = 3.82VVTKK396 pKa = 10.78GSFSFSNFTRR406 pKa = 11.84EE407 pKa = 4.33RR408 pKa = 11.84RR409 pKa = 11.84FHH411 pKa = 6.97LNFDD415 pKa = 3.99GLATNVNQIVTDD427 pKa = 3.8DD428 pKa = 3.72TDD430 pKa = 5.08LIGPVQEE437 pKa = 4.28AA438 pKa = 3.48
Molecular weight: 47.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
990 |
195 |
438 |
330.0 |
36.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.475 ± 0.279 | 2.02 ± 0.791 |
5.859 ± 0.765 | 4.444 ± 1.522 |
4.141 ± 0.682 | 7.879 ± 1.437 |
3.838 ± 1.025 | 4.646 ± 0.721 |
5.152 ± 1.068 | 6.97 ± 0.504 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.313 ± 0.277 | 4.444 ± 0.489 |
5.556 ± 0.818 | 4.04 ± 0.511 |
7.677 ± 0.701 | 6.97 ± 0.998 |
6.465 ± 1.262 | 6.162 ± 0.234 |
1.818 ± 0.687 | 3.131 ± 0.152 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
