
Murine roseolovirus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Roseolovirus; unclassified Roseolovirus
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
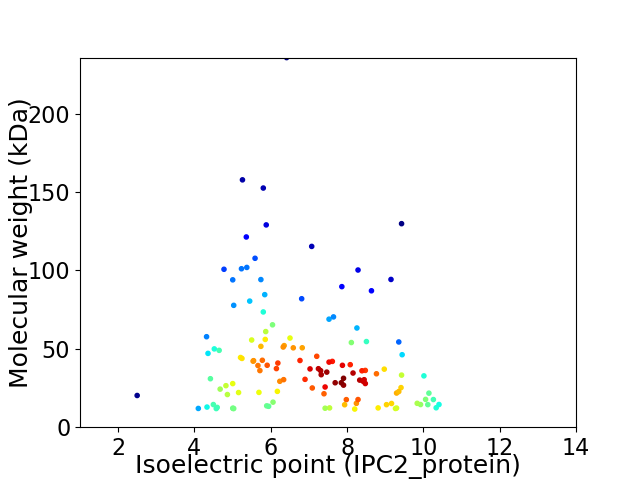
Virtual 2D-PAGE plot for 128 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1P8VIR2|A0A1P8VIR2_9BETA Uncharacterized protein OS=Murine roseolovirus OX=1940555 GN=ORF10 PE=4 SV=1
MM1 pKa = 7.98CYY3 pKa = 10.57HH4 pKa = 7.15MLPSHH9 pKa = 5.53QQEE12 pKa = 3.69EE13 pKa = 4.7MEE15 pKa = 4.54NNEE18 pKa = 4.15SSDD21 pKa = 3.77TVEE24 pKa = 4.38ATPSDD29 pKa = 3.84TDD31 pKa = 3.9QVSLPHH37 pKa = 6.71GNPDD41 pKa = 3.35SPTDD45 pKa = 3.5GTAYY49 pKa = 9.8PDD51 pKa = 3.84LHH53 pKa = 7.12DD54 pKa = 5.58LPPPIIDD61 pKa = 5.4PITGTVVLTLLEE73 pKa = 4.51MPAPPHH79 pKa = 6.87PPTPPPIPPRR89 pKa = 11.84TYY91 pKa = 9.93KK92 pKa = 10.62PMLITHH98 pKa = 6.54PEE100 pKa = 3.86EE101 pKa = 6.56DD102 pKa = 3.93EE103 pKa = 4.27EE104 pKa = 4.65QLDD107 pKa = 3.78RR108 pKa = 6.35
MM1 pKa = 7.98CYY3 pKa = 10.57HH4 pKa = 7.15MLPSHH9 pKa = 5.53QQEE12 pKa = 3.69EE13 pKa = 4.7MEE15 pKa = 4.54NNEE18 pKa = 4.15SSDD21 pKa = 3.77TVEE24 pKa = 4.38ATPSDD29 pKa = 3.84TDD31 pKa = 3.9QVSLPHH37 pKa = 6.71GNPDD41 pKa = 3.35SPTDD45 pKa = 3.5GTAYY49 pKa = 9.8PDD51 pKa = 3.84LHH53 pKa = 7.12DD54 pKa = 5.58LPPPIIDD61 pKa = 5.4PITGTVVLTLLEE73 pKa = 4.51MPAPPHH79 pKa = 6.87PPTPPPIPPRR89 pKa = 11.84TYY91 pKa = 9.93KK92 pKa = 10.62PMLITHH98 pKa = 6.54PEE100 pKa = 3.86EE101 pKa = 6.56DD102 pKa = 3.93EE103 pKa = 4.27EE104 pKa = 4.65QLDD107 pKa = 3.78RR108 pKa = 6.35
Molecular weight: 11.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8VIZ9|A0A1P8VIZ9_9BETA Uncharacterized protein OS=Murine roseolovirus OX=1940555 GN=ORF111 PE=4 SV=1
MM1 pKa = 7.82IDD3 pKa = 3.29TEE5 pKa = 4.66QFLEE9 pKa = 4.47CDD11 pKa = 3.46HH12 pKa = 6.23GHH14 pKa = 6.4CHH16 pKa = 6.68TIAISGNSADD26 pKa = 4.56LRR28 pKa = 11.84PLVAVRR34 pKa = 11.84FSFFVFRR41 pKa = 11.84ALFCHH46 pKa = 6.61HH47 pKa = 7.33GDD49 pKa = 3.91GPLPSPPPSPWQPLLPLRR67 pKa = 11.84AARR70 pKa = 11.84VTTVVLIRR78 pKa = 11.84TPPVRR83 pKa = 11.84LGCPLTGVAMIKK95 pKa = 8.37TAPVSAAAPLPCVSVSVSACRR116 pKa = 11.84CRR118 pKa = 11.84VSCVPSLGVSHH129 pKa = 7.21FSSSYY134 pKa = 10.42SSPLGPSPTPHH145 pKa = 7.31PSLTAAATSARR156 pKa = 11.84QTLAPSSHH164 pKa = 5.41GRR166 pKa = 11.84EE167 pKa = 3.98EE168 pKa = 3.83QTLDD172 pKa = 3.36FLQSRR177 pKa = 11.84DD178 pKa = 3.31RR179 pKa = 11.84DD180 pKa = 3.8FLTKK184 pKa = 10.3KK185 pKa = 10.08RR186 pKa = 11.84DD187 pKa = 3.56RR188 pKa = 11.84FQIWYY193 pKa = 9.4LPLNGSCCRR202 pKa = 11.84RR203 pKa = 11.84CCWTLCLTAKK213 pKa = 10.29PSVFSPSRR221 pKa = 11.84GCLWSAAVAAPPRR234 pKa = 11.84SSPTPPGALAAAPPAEE250 pKa = 4.74PGNRR254 pKa = 11.84SCGGPPPTNSGRR266 pKa = 11.84TSGPRR271 pKa = 11.84MIGCTTSRR279 pKa = 11.84LRR281 pKa = 11.84TGSAPCAPPCYY292 pKa = 9.88GVGASWPPPGPPGGACTWNRR312 pKa = 11.84TPTASAWPSTRR323 pKa = 11.84CPSPVSVSAGRR334 pKa = 11.84PSPGPPTRR342 pKa = 11.84WNLSVRR348 pKa = 11.84MLFPCLPLPKK358 pKa = 9.95FPFPPHH364 pKa = 6.94PSPTPLTSFVSACLSSAVSHH384 pKa = 5.98VVLSSGVVPLFGVSRR399 pKa = 11.84DD400 pKa = 3.66YY401 pKa = 11.37VPPRR405 pKa = 11.84RR406 pKa = 11.84VGGEE410 pKa = 3.53RR411 pKa = 11.84LVWFFCRR418 pKa = 11.84KK419 pKa = 8.39EE420 pKa = 3.96KK421 pKa = 9.55KK422 pKa = 8.01TCFSILSRR430 pKa = 11.84RR431 pKa = 11.84PLLSLLAPSRR441 pKa = 11.84SPLSVLCSPLAPSRR455 pKa = 11.84LPVRR459 pKa = 11.84IFPLSSPSLRR469 pKa = 11.84VPHH472 pKa = 6.34PHH474 pKa = 7.16PLSCLSLPRR483 pKa = 11.84SEE485 pKa = 4.46RR486 pKa = 11.84APIQVLWGHH495 pKa = 6.17RR496 pKa = 11.84VFLPGGLHH504 pKa = 6.18RR505 pKa = 11.84ASPRR509 pKa = 11.84AA510 pKa = 3.35
MM1 pKa = 7.82IDD3 pKa = 3.29TEE5 pKa = 4.66QFLEE9 pKa = 4.47CDD11 pKa = 3.46HH12 pKa = 6.23GHH14 pKa = 6.4CHH16 pKa = 6.68TIAISGNSADD26 pKa = 4.56LRR28 pKa = 11.84PLVAVRR34 pKa = 11.84FSFFVFRR41 pKa = 11.84ALFCHH46 pKa = 6.61HH47 pKa = 7.33GDD49 pKa = 3.91GPLPSPPPSPWQPLLPLRR67 pKa = 11.84AARR70 pKa = 11.84VTTVVLIRR78 pKa = 11.84TPPVRR83 pKa = 11.84LGCPLTGVAMIKK95 pKa = 8.37TAPVSAAAPLPCVSVSVSACRR116 pKa = 11.84CRR118 pKa = 11.84VSCVPSLGVSHH129 pKa = 7.21FSSSYY134 pKa = 10.42SSPLGPSPTPHH145 pKa = 7.31PSLTAAATSARR156 pKa = 11.84QTLAPSSHH164 pKa = 5.41GRR166 pKa = 11.84EE167 pKa = 3.98EE168 pKa = 3.83QTLDD172 pKa = 3.36FLQSRR177 pKa = 11.84DD178 pKa = 3.31RR179 pKa = 11.84DD180 pKa = 3.8FLTKK184 pKa = 10.3KK185 pKa = 10.08RR186 pKa = 11.84DD187 pKa = 3.56RR188 pKa = 11.84FQIWYY193 pKa = 9.4LPLNGSCCRR202 pKa = 11.84RR203 pKa = 11.84CCWTLCLTAKK213 pKa = 10.29PSVFSPSRR221 pKa = 11.84GCLWSAAVAAPPRR234 pKa = 11.84SSPTPPGALAAAPPAEE250 pKa = 4.74PGNRR254 pKa = 11.84SCGGPPPTNSGRR266 pKa = 11.84TSGPRR271 pKa = 11.84MIGCTTSRR279 pKa = 11.84LRR281 pKa = 11.84TGSAPCAPPCYY292 pKa = 9.88GVGASWPPPGPPGGACTWNRR312 pKa = 11.84TPTASAWPSTRR323 pKa = 11.84CPSPVSVSAGRR334 pKa = 11.84PSPGPPTRR342 pKa = 11.84WNLSVRR348 pKa = 11.84MLFPCLPLPKK358 pKa = 9.95FPFPPHH364 pKa = 6.94PSPTPLTSFVSACLSSAVSHH384 pKa = 5.98VVLSSGVVPLFGVSRR399 pKa = 11.84DD400 pKa = 3.66YY401 pKa = 11.37VPPRR405 pKa = 11.84RR406 pKa = 11.84VGGEE410 pKa = 3.53RR411 pKa = 11.84LVWFFCRR418 pKa = 11.84KK419 pKa = 8.39EE420 pKa = 3.96KK421 pKa = 9.55KK422 pKa = 8.01TCFSILSRR430 pKa = 11.84RR431 pKa = 11.84PLLSLLAPSRR441 pKa = 11.84SPLSVLCSPLAPSRR455 pKa = 11.84LPVRR459 pKa = 11.84IFPLSSPSLRR469 pKa = 11.84VPHH472 pKa = 6.34PHH474 pKa = 7.16PLSCLSLPRR483 pKa = 11.84SEE485 pKa = 4.46RR486 pKa = 11.84APIQVLWGHH495 pKa = 6.17RR496 pKa = 11.84VFLPGGLHH504 pKa = 6.18RR505 pKa = 11.84ASPRR509 pKa = 11.84AA510 pKa = 3.35
Molecular weight: 54.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
50423 |
105 |
2011 |
393.9 |
45.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.012 ± 0.237 | 2.303 ± 0.127 |
5.503 ± 0.186 | 5.648 ± 0.196 |
5.317 ± 0.195 | 4.155 ± 0.295 |
2.366 ± 0.097 | 8.611 ± 0.343 |
7.126 ± 0.275 | 9.101 ± 0.227 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.39 ± 0.092 | 6.715 ± 0.31 |
5.091 ± 0.408 | 3.407 ± 0.153 |
4.107 ± 0.258 | 8.012 ± 0.422 |
5.948 ± 0.177 | 5.053 ± 0.149 |
0.837 ± 0.056 | 4.298 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
