
Odonata-associated circular virus-18
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins
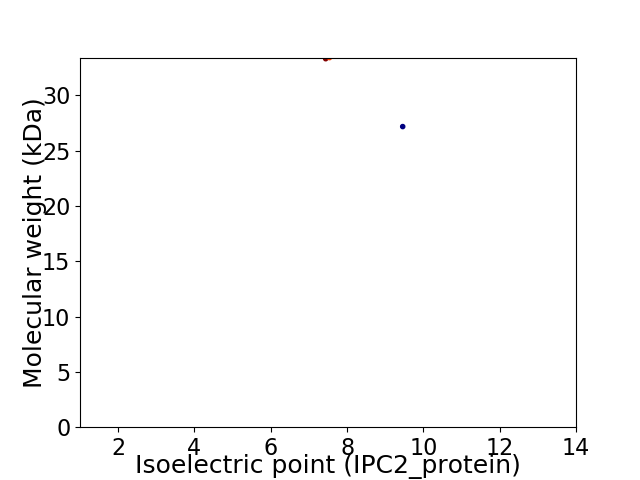
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
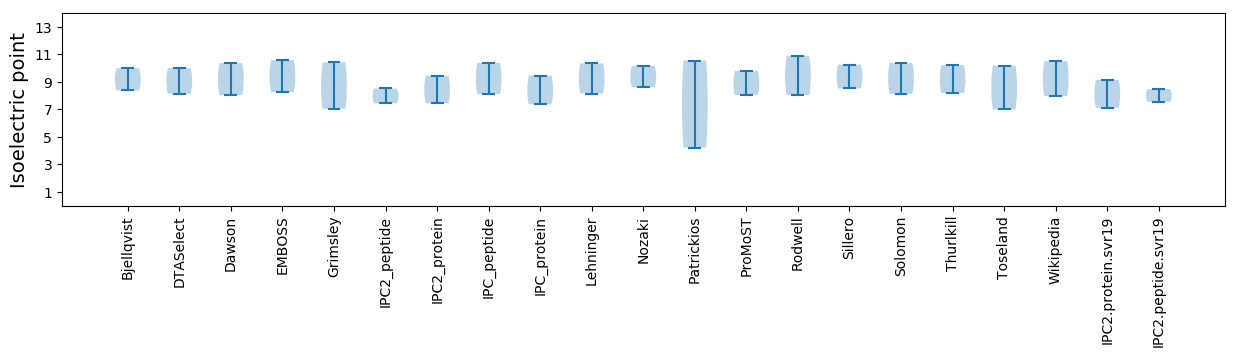
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI18|A0A0B4UI18_9VIRU Putative capsid protein OS=Odonata-associated circular virus-18 OX=1592118 PE=4 SV=1
MM1 pKa = 7.47SKK3 pKa = 7.24TTRR6 pKa = 11.84FCFTKK11 pKa = 10.41NNYY14 pKa = 9.58SEE16 pKa = 4.41LHH18 pKa = 5.35VDD20 pKa = 3.23ILKK23 pKa = 10.57SDD25 pKa = 3.6LTSTFKK31 pKa = 10.65YY32 pKa = 7.72WCFGIEE38 pKa = 4.14VGEE41 pKa = 4.34SGTPHH46 pKa = 5.66LQGYY50 pKa = 8.41FEE52 pKa = 5.0FPNKK56 pKa = 10.17NPLRR60 pKa = 11.84ISGAQNKK67 pKa = 9.37LNNIGLEE74 pKa = 4.14GCAIFPAKK82 pKa = 8.95GTAEE86 pKa = 3.94QNITYY91 pKa = 9.78CSKK94 pKa = 10.85DD95 pKa = 3.1GSFFEE100 pKa = 4.87GGEE103 pKa = 4.04RR104 pKa = 11.84PRR106 pKa = 11.84GQGKK110 pKa = 7.81RR111 pKa = 11.84TDD113 pKa = 3.84LDD115 pKa = 4.11SVCEE119 pKa = 4.33LIKK122 pKa = 10.95SGANMTEE129 pKa = 4.21VANQFPSQVVKK140 pKa = 9.84FHH142 pKa = 6.99SGLQFLMNVTITPRR156 pKa = 11.84RR157 pKa = 11.84FKK159 pKa = 11.03TEE161 pKa = 3.88VWWLWGPTGSGKK173 pKa = 10.09SRR175 pKa = 11.84YY176 pKa = 9.56AWDD179 pKa = 4.1QYY181 pKa = 10.06PSSYY185 pKa = 10.11MKK187 pKa = 10.42EE188 pKa = 4.6CSHH191 pKa = 6.34KK192 pKa = 9.73WWDD195 pKa = 3.71GYY197 pKa = 9.37TGQDD201 pKa = 3.48VVIMDD206 pKa = 4.59DD207 pKa = 3.82FRR209 pKa = 11.84PSKK212 pKa = 9.96EE213 pKa = 4.09LPFSFILNLFDD224 pKa = 5.56RR225 pKa = 11.84YY226 pKa = 9.75PLSVQVKK233 pKa = 8.62GGMAQFVSKK242 pKa = 9.31TIYY245 pKa = 9.1VTTPLSPEE253 pKa = 4.02DD254 pKa = 3.82TCNHH258 pKa = 6.23LEE260 pKa = 4.11WLGPEE265 pKa = 3.91QKK267 pKa = 10.32EE268 pKa = 4.01QFLRR272 pKa = 11.84RR273 pKa = 11.84IDD275 pKa = 3.87HH276 pKa = 6.54IVRR279 pKa = 11.84FPQMMTNYY287 pKa = 10.28LARR290 pKa = 5.36
MM1 pKa = 7.47SKK3 pKa = 7.24TTRR6 pKa = 11.84FCFTKK11 pKa = 10.41NNYY14 pKa = 9.58SEE16 pKa = 4.41LHH18 pKa = 5.35VDD20 pKa = 3.23ILKK23 pKa = 10.57SDD25 pKa = 3.6LTSTFKK31 pKa = 10.65YY32 pKa = 7.72WCFGIEE38 pKa = 4.14VGEE41 pKa = 4.34SGTPHH46 pKa = 5.66LQGYY50 pKa = 8.41FEE52 pKa = 5.0FPNKK56 pKa = 10.17NPLRR60 pKa = 11.84ISGAQNKK67 pKa = 9.37LNNIGLEE74 pKa = 4.14GCAIFPAKK82 pKa = 8.95GTAEE86 pKa = 3.94QNITYY91 pKa = 9.78CSKK94 pKa = 10.85DD95 pKa = 3.1GSFFEE100 pKa = 4.87GGEE103 pKa = 4.04RR104 pKa = 11.84PRR106 pKa = 11.84GQGKK110 pKa = 7.81RR111 pKa = 11.84TDD113 pKa = 3.84LDD115 pKa = 4.11SVCEE119 pKa = 4.33LIKK122 pKa = 10.95SGANMTEE129 pKa = 4.21VANQFPSQVVKK140 pKa = 9.84FHH142 pKa = 6.99SGLQFLMNVTITPRR156 pKa = 11.84RR157 pKa = 11.84FKK159 pKa = 11.03TEE161 pKa = 3.88VWWLWGPTGSGKK173 pKa = 10.09SRR175 pKa = 11.84YY176 pKa = 9.56AWDD179 pKa = 4.1QYY181 pKa = 10.06PSSYY185 pKa = 10.11MKK187 pKa = 10.42EE188 pKa = 4.6CSHH191 pKa = 6.34KK192 pKa = 9.73WWDD195 pKa = 3.71GYY197 pKa = 9.37TGQDD201 pKa = 3.48VVIMDD206 pKa = 4.59DD207 pKa = 3.82FRR209 pKa = 11.84PSKK212 pKa = 9.96EE213 pKa = 4.09LPFSFILNLFDD224 pKa = 5.56RR225 pKa = 11.84YY226 pKa = 9.75PLSVQVKK233 pKa = 8.62GGMAQFVSKK242 pKa = 9.31TIYY245 pKa = 9.1VTTPLSPEE253 pKa = 4.02DD254 pKa = 3.82TCNHH258 pKa = 6.23LEE260 pKa = 4.11WLGPEE265 pKa = 3.91QKK267 pKa = 10.32EE268 pKa = 4.01QFLRR272 pKa = 11.84RR273 pKa = 11.84IDD275 pKa = 3.87HH276 pKa = 6.54IVRR279 pKa = 11.84FPQMMTNYY287 pKa = 10.28LARR290 pKa = 5.36
Molecular weight: 33.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI18|A0A0B4UI18_9VIRU Putative capsid protein OS=Odonata-associated circular virus-18 OX=1592118 PE=4 SV=1
MM1 pKa = 8.13AYY3 pKa = 10.02RR4 pKa = 11.84KK5 pKa = 10.18KK6 pKa = 9.9KK7 pKa = 6.98TFKK10 pKa = 10.17RR11 pKa = 11.84KK12 pKa = 9.53RR13 pKa = 11.84SFKK16 pKa = 10.39RR17 pKa = 11.84PSTRR21 pKa = 11.84VSKK24 pKa = 10.32KK25 pKa = 8.39VKK27 pKa = 10.19RR28 pKa = 11.84FVKK31 pKa = 10.21KK32 pKa = 10.53YY33 pKa = 9.54VDD35 pKa = 3.42RR36 pKa = 11.84QIEE39 pKa = 4.29DD40 pKa = 3.22KK41 pKa = 10.77RR42 pKa = 11.84VQTTQTEE49 pKa = 4.52VTYY52 pKa = 11.39SNTLTNAQVFSLMPSIGQGVDD73 pKa = 2.95DD74 pKa = 4.65SSRR77 pKa = 11.84TGNVVRR83 pKa = 11.84LRR85 pKa = 11.84KK86 pKa = 9.87AILRR90 pKa = 11.84LSASAFDD97 pKa = 4.53NNIHH101 pKa = 6.12AHH103 pKa = 5.88WNDD106 pKa = 3.05LYY108 pKa = 10.75IFKK111 pKa = 10.36KK112 pKa = 9.53QAGIVAPTAAEE123 pKa = 3.97MNLFLDD129 pKa = 4.1NGSTAVAYY137 pKa = 9.95AGQPLGGLRR146 pKa = 11.84PVNPFTFIKK155 pKa = 10.1KK156 pKa = 8.17LHH158 pKa = 6.47KK159 pKa = 8.53RR160 pKa = 11.84TMMASVQSGGASGSAIGSVKK180 pKa = 10.44PCFTMTKK187 pKa = 10.14DD188 pKa = 2.78ITSFFKK194 pKa = 10.85KK195 pKa = 9.3RR196 pKa = 11.84WVYY199 pKa = 10.94EE200 pKa = 3.97DD201 pKa = 3.76STLSTPNNDD210 pKa = 2.82NVFIGMGGSDD220 pKa = 3.68MTGQIAANYY229 pKa = 9.32GEE231 pKa = 4.24YY232 pKa = 10.15SYY234 pKa = 11.74SVDD237 pKa = 4.8FFYY240 pKa = 11.35EE241 pKa = 4.08DD242 pKa = 3.0AA243 pKa = 5.45
MM1 pKa = 8.13AYY3 pKa = 10.02RR4 pKa = 11.84KK5 pKa = 10.18KK6 pKa = 9.9KK7 pKa = 6.98TFKK10 pKa = 10.17RR11 pKa = 11.84KK12 pKa = 9.53RR13 pKa = 11.84SFKK16 pKa = 10.39RR17 pKa = 11.84PSTRR21 pKa = 11.84VSKK24 pKa = 10.32KK25 pKa = 8.39VKK27 pKa = 10.19RR28 pKa = 11.84FVKK31 pKa = 10.21KK32 pKa = 10.53YY33 pKa = 9.54VDD35 pKa = 3.42RR36 pKa = 11.84QIEE39 pKa = 4.29DD40 pKa = 3.22KK41 pKa = 10.77RR42 pKa = 11.84VQTTQTEE49 pKa = 4.52VTYY52 pKa = 11.39SNTLTNAQVFSLMPSIGQGVDD73 pKa = 2.95DD74 pKa = 4.65SSRR77 pKa = 11.84TGNVVRR83 pKa = 11.84LRR85 pKa = 11.84KK86 pKa = 9.87AILRR90 pKa = 11.84LSASAFDD97 pKa = 4.53NNIHH101 pKa = 6.12AHH103 pKa = 5.88WNDD106 pKa = 3.05LYY108 pKa = 10.75IFKK111 pKa = 10.36KK112 pKa = 9.53QAGIVAPTAAEE123 pKa = 3.97MNLFLDD129 pKa = 4.1NGSTAVAYY137 pKa = 9.95AGQPLGGLRR146 pKa = 11.84PVNPFTFIKK155 pKa = 10.1KK156 pKa = 8.17LHH158 pKa = 6.47KK159 pKa = 8.53RR160 pKa = 11.84TMMASVQSGGASGSAIGSVKK180 pKa = 10.44PCFTMTKK187 pKa = 10.14DD188 pKa = 2.78ITSFFKK194 pKa = 10.85KK195 pKa = 9.3RR196 pKa = 11.84WVYY199 pKa = 10.94EE200 pKa = 3.97DD201 pKa = 3.76STLSTPNNDD210 pKa = 2.82NVFIGMGGSDD220 pKa = 3.68MTGQIAANYY229 pKa = 9.32GEE231 pKa = 4.24YY232 pKa = 10.15SYY234 pKa = 11.74SVDD237 pKa = 4.8FFYY240 pKa = 11.35EE241 pKa = 4.08DD242 pKa = 3.0AA243 pKa = 5.45
Molecular weight: 27.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
533 |
243 |
290 |
266.5 |
30.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.253 ± 1.809 | 1.501 ± 0.768 |
4.878 ± 0.333 | 4.315 ± 1.302 |
6.567 ± 0.278 | 7.505 ± 0.359 |
1.689 ± 0.32 | 4.503 ± 0.017 |
7.692 ± 0.96 | 6.191 ± 0.884 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.002 ± 0.205 | 5.066 ± 0.2 |
4.503 ± 0.854 | 4.315 ± 0.431 |
5.441 ± 0.516 | 8.255 ± 0.563 |
7.317 ± 0.354 | 6.191 ± 0.858 |
1.876 ± 0.743 | 3.94 ± 0.124 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
