
Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
Taxonomy: cellular organisms; Bacteria; Thermotogae; Thermotogae; Thermotogales; Thermotogaceae; Thermotoga; Thermotoga maritima
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
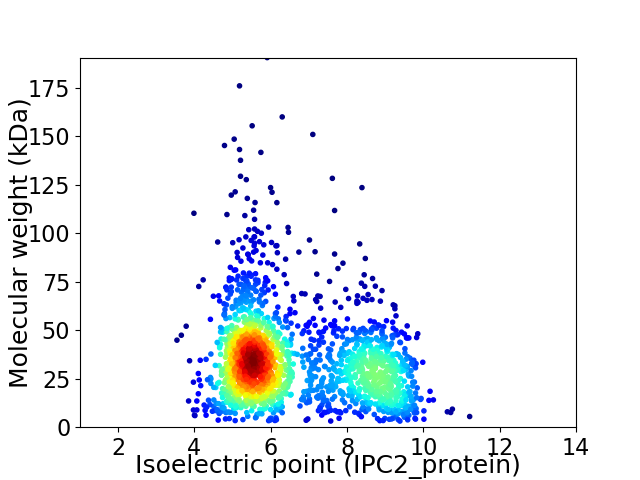
Virtual 2D-PAGE plot for 1852 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9X2G0|Q9X2G0_THEMA Uncharacterized protein OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=TM_1844 PE=4 SV=1
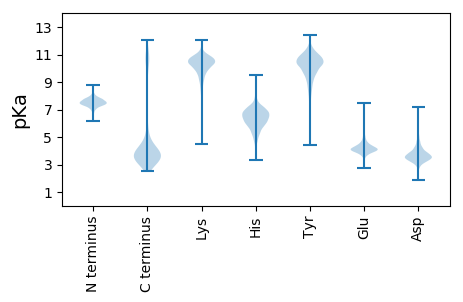
MM1 pKa = 7.57KK2 pKa = 10.58KK3 pKa = 10.37LFLVLAVLSLVLYY16 pKa = 10.2GCLRR20 pKa = 11.84TPPVVKK26 pKa = 10.79DD27 pKa = 3.35MVTTDD32 pKa = 3.79GSLSDD37 pKa = 3.4WGSKK41 pKa = 9.56IKK43 pKa = 10.55EE44 pKa = 4.12DD45 pKa = 4.81AADD48 pKa = 3.95DD49 pKa = 4.41SKK51 pKa = 11.14WGADD55 pKa = 3.51NEE57 pKa = 4.19LLKK60 pKa = 11.11AGLLFDD66 pKa = 4.58GANIYY71 pKa = 10.04IAGEE75 pKa = 3.98YY76 pKa = 10.24VIGGDD81 pKa = 3.55GNVNVFLVLIDD92 pKa = 3.81LVGVTGAQTVSYY104 pKa = 10.07TGLNDD109 pKa = 3.39GNRR112 pKa = 11.84NFTNPNGDD120 pKa = 2.72IDD122 pKa = 6.23LIVEE126 pKa = 4.26VNQSNEE132 pKa = 3.84YY133 pKa = 9.94KK134 pKa = 9.9VWRR137 pKa = 11.84VSQDD141 pKa = 2.66GSLTDD146 pKa = 3.17ITDD149 pKa = 3.2QVTAQFGTGSTIAEE163 pKa = 4.1LKK165 pKa = 10.53IPISEE170 pKa = 4.04QVNQVKK176 pKa = 10.15AVFAISGGTGGGNQWVGDD194 pKa = 4.5FYY196 pKa = 10.99PNQPDD201 pKa = 3.32HH202 pKa = 7.62DD203 pKa = 4.27SAEE206 pKa = 4.33NGGIPQPAPVVNFVVCDD223 pKa = 3.56KK224 pKa = 10.39EE225 pKa = 5.15GNVSEE230 pKa = 5.27EE231 pKa = 4.1INQTSEE237 pKa = 4.14PEE239 pKa = 3.97QPTEE243 pKa = 3.92SVIVVDD249 pKa = 5.33GDD251 pKa = 3.64LSDD254 pKa = 4.58FGTPVATSTNPGGGNGANLYY274 pKa = 9.77RR275 pKa = 11.84LYY277 pKa = 9.86VTYY280 pKa = 10.57DD281 pKa = 2.87ATYY284 pKa = 10.37LYY286 pKa = 10.33IGFDD290 pKa = 3.85TQNSGSWDD298 pKa = 3.31VAYY301 pKa = 10.31GIGIDD306 pKa = 4.14TKK308 pKa = 10.18PSGYY312 pKa = 9.25TGDD315 pKa = 3.35SDD317 pKa = 3.03AWGRR321 pKa = 11.84KK322 pKa = 7.76ISFGADD328 pKa = 3.06YY329 pKa = 11.09AVDD332 pKa = 3.57YY333 pKa = 10.62EE334 pKa = 5.16IYY336 pKa = 9.77FWWAGGSGLDD346 pKa = 4.54SNNFCEE352 pKa = 5.2WNGLDD357 pKa = 3.08WNYY360 pKa = 11.04RR361 pKa = 11.84SIADD365 pKa = 3.3AGGTFDD371 pKa = 3.69YY372 pKa = 11.09TGDD375 pKa = 3.74TSSGLQTMEE384 pKa = 4.46IAIPWSAIGGKK395 pKa = 9.28QDD397 pKa = 2.76VALIVWITGGSGSAVDD413 pKa = 5.69SIPDD417 pKa = 3.98DD418 pKa = 5.22DD419 pKa = 4.28STIDD423 pKa = 6.1NADD426 pKa = 3.13EE427 pKa = 4.36WNDD430 pKa = 3.09SDD432 pKa = 4.81TFTNLYY438 pKa = 9.9LLQVNN443 pKa = 4.57
MM1 pKa = 7.57KK2 pKa = 10.58KK3 pKa = 10.37LFLVLAVLSLVLYY16 pKa = 10.2GCLRR20 pKa = 11.84TPPVVKK26 pKa = 10.79DD27 pKa = 3.35MVTTDD32 pKa = 3.79GSLSDD37 pKa = 3.4WGSKK41 pKa = 9.56IKK43 pKa = 10.55EE44 pKa = 4.12DD45 pKa = 4.81AADD48 pKa = 3.95DD49 pKa = 4.41SKK51 pKa = 11.14WGADD55 pKa = 3.51NEE57 pKa = 4.19LLKK60 pKa = 11.11AGLLFDD66 pKa = 4.58GANIYY71 pKa = 10.04IAGEE75 pKa = 3.98YY76 pKa = 10.24VIGGDD81 pKa = 3.55GNVNVFLVLIDD92 pKa = 3.81LVGVTGAQTVSYY104 pKa = 10.07TGLNDD109 pKa = 3.39GNRR112 pKa = 11.84NFTNPNGDD120 pKa = 2.72IDD122 pKa = 6.23LIVEE126 pKa = 4.26VNQSNEE132 pKa = 3.84YY133 pKa = 9.94KK134 pKa = 9.9VWRR137 pKa = 11.84VSQDD141 pKa = 2.66GSLTDD146 pKa = 3.17ITDD149 pKa = 3.2QVTAQFGTGSTIAEE163 pKa = 4.1LKK165 pKa = 10.53IPISEE170 pKa = 4.04QVNQVKK176 pKa = 10.15AVFAISGGTGGGNQWVGDD194 pKa = 4.5FYY196 pKa = 10.99PNQPDD201 pKa = 3.32HH202 pKa = 7.62DD203 pKa = 4.27SAEE206 pKa = 4.33NGGIPQPAPVVNFVVCDD223 pKa = 3.56KK224 pKa = 10.39EE225 pKa = 5.15GNVSEE230 pKa = 5.27EE231 pKa = 4.1INQTSEE237 pKa = 4.14PEE239 pKa = 3.97QPTEE243 pKa = 3.92SVIVVDD249 pKa = 5.33GDD251 pKa = 3.64LSDD254 pKa = 4.58FGTPVATSTNPGGGNGANLYY274 pKa = 9.77RR275 pKa = 11.84LYY277 pKa = 9.86VTYY280 pKa = 10.57DD281 pKa = 2.87ATYY284 pKa = 10.37LYY286 pKa = 10.33IGFDD290 pKa = 3.85TQNSGSWDD298 pKa = 3.31VAYY301 pKa = 10.31GIGIDD306 pKa = 4.14TKK308 pKa = 10.18PSGYY312 pKa = 9.25TGDD315 pKa = 3.35SDD317 pKa = 3.03AWGRR321 pKa = 11.84KK322 pKa = 7.76ISFGADD328 pKa = 3.06YY329 pKa = 11.09AVDD332 pKa = 3.57YY333 pKa = 10.62EE334 pKa = 5.16IYY336 pKa = 9.77FWWAGGSGLDD346 pKa = 4.54SNNFCEE352 pKa = 5.2WNGLDD357 pKa = 3.08WNYY360 pKa = 11.04RR361 pKa = 11.84SIADD365 pKa = 3.3AGGTFDD371 pKa = 3.69YY372 pKa = 11.09TGDD375 pKa = 3.74TSSGLQTMEE384 pKa = 4.46IAIPWSAIGGKK395 pKa = 9.28QDD397 pKa = 2.76VALIVWITGGSGSAVDD413 pKa = 5.69SIPDD417 pKa = 3.98DD418 pKa = 5.22DD419 pKa = 4.28STIDD423 pKa = 6.1NADD426 pKa = 3.13EE427 pKa = 4.36WNDD430 pKa = 3.09SDD432 pKa = 4.81TFTNLYY438 pKa = 9.9LLQVNN443 pKa = 4.57
Molecular weight: 47.42 kDa
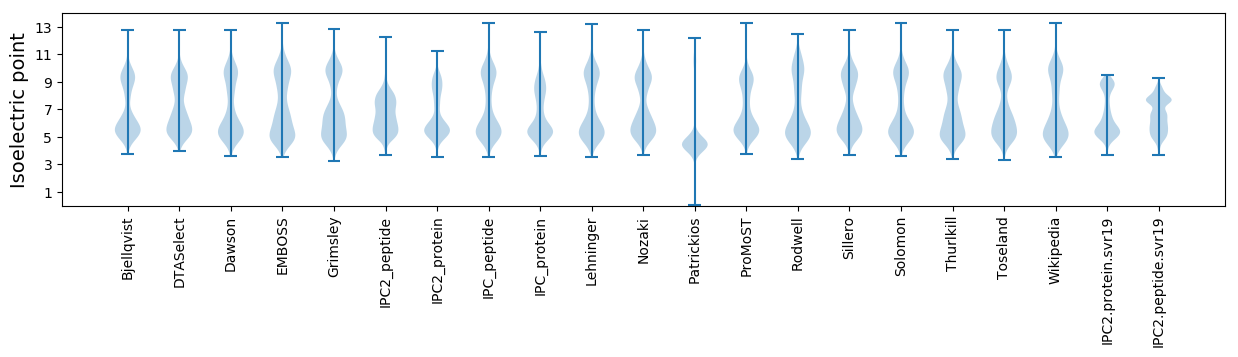
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9WXM0|Q9WXM0_THEMA DNA helicase putative OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=TM_0005 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.16QPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.66RR12 pKa = 11.84KK13 pKa = 8.13RR14 pKa = 11.84THH16 pKa = 6.11GFLARR21 pKa = 11.84KK22 pKa = 6.6RR23 pKa = 11.84TPGGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 9.72NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.65GRR39 pKa = 11.84WRR41 pKa = 11.84LTVV44 pKa = 3.0
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.16QPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.66RR12 pKa = 11.84KK13 pKa = 8.13RR14 pKa = 11.84THH16 pKa = 6.11GFLARR21 pKa = 11.84KK22 pKa = 6.6RR23 pKa = 11.84TPGGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 9.72NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.65GRR39 pKa = 11.84WRR41 pKa = 11.84LTVV44 pKa = 3.0
Molecular weight: 5.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
582801 |
30 |
1690 |
314.7 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.849 ± 0.051 | 0.708 ± 0.016 |
4.966 ± 0.036 | 8.924 ± 0.065 |
5.195 ± 0.044 | 6.908 ± 0.045 |
1.585 ± 0.019 | 7.185 ± 0.037 |
7.617 ± 0.051 | 10.039 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.023 | 3.61 ± 0.031 |
3.993 ± 0.033 | 2.013 ± 0.026 |
5.536 ± 0.048 | 5.651 ± 0.045 |
4.526 ± 0.031 | 8.61 ± 0.052 |
1.102 ± 0.022 | 3.58 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
