
Gammapapillomavirus 25
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
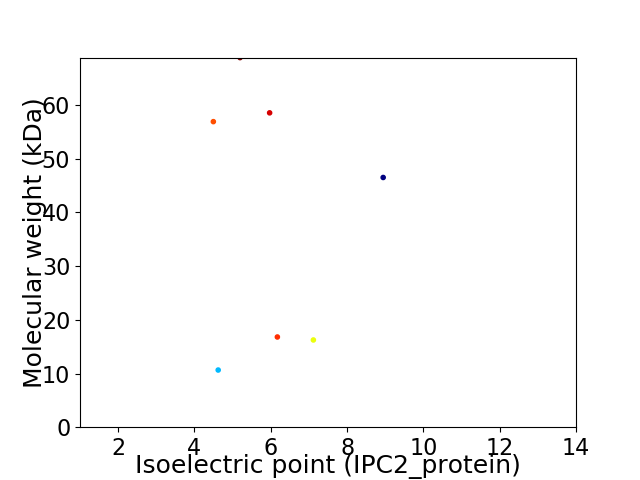
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2AMD9|A0A2D2AMD9_9PAPI Minor capsid protein L2 OS=Gammapapillomavirus 25 OX=1961682 GN=L2 PE=3 SV=1
MM1 pKa = 7.67IGPSPNNDD9 pKa = 4.03DD10 pKa = 5.03IEE12 pKa = 6.57LDD14 pKa = 3.5LHH16 pKa = 7.27DD17 pKa = 5.77LVLPSNLISEE27 pKa = 4.91EE28 pKa = 4.12SLSPDD33 pKa = 2.79VDD35 pKa = 3.83PEE37 pKa = 4.19EE38 pKa = 5.06EE39 pKa = 4.16EE40 pKa = 3.95QLPFAVEE47 pKa = 4.53TSCKK51 pKa = 10.04SCGAGVRR58 pKa = 11.84FTVCATHH65 pKa = 6.78AAIRR69 pKa = 11.84TLQILLLQEE78 pKa = 4.58LSLICLRR85 pKa = 11.84CSRR88 pKa = 11.84SLIQHH93 pKa = 6.13GRR95 pKa = 11.84PHH97 pKa = 6.63
MM1 pKa = 7.67IGPSPNNDD9 pKa = 4.03DD10 pKa = 5.03IEE12 pKa = 6.57LDD14 pKa = 3.5LHH16 pKa = 7.27DD17 pKa = 5.77LVLPSNLISEE27 pKa = 4.91EE28 pKa = 4.12SLSPDD33 pKa = 2.79VDD35 pKa = 3.83PEE37 pKa = 4.19EE38 pKa = 5.06EE39 pKa = 4.16EE40 pKa = 3.95QLPFAVEE47 pKa = 4.53TSCKK51 pKa = 10.04SCGAGVRR58 pKa = 11.84FTVCATHH65 pKa = 6.78AAIRR69 pKa = 11.84TLQILLLQEE78 pKa = 4.58LSLICLRR85 pKa = 11.84CSRR88 pKa = 11.84SLIQHH93 pKa = 6.13GRR95 pKa = 11.84PHH97 pKa = 6.63
Molecular weight: 10.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AMB7|A0A2D2AMB7_9PAPI E4 OS=Gammapapillomavirus 25 OX=1961682 GN=E4 PE=4 SV=1
MM1 pKa = 7.0NQADD5 pKa = 3.68LTARR9 pKa = 11.84FDD11 pKa = 3.97ALQEE15 pKa = 4.17TLMTLYY21 pKa = 10.29EE22 pKa = 4.17RR23 pKa = 11.84NARR26 pKa = 11.84DD27 pKa = 3.6LEE29 pKa = 4.26SQILHH34 pKa = 6.42WEE36 pKa = 4.49TVRR39 pKa = 11.84KK40 pKa = 9.93QYY42 pKa = 9.59VTMYY46 pKa = 9.06YY47 pKa = 10.43ARR49 pKa = 11.84KK50 pKa = 9.55EE51 pKa = 4.26GFRR54 pKa = 11.84SLGMQPLPALTVSEE68 pKa = 4.51YY69 pKa = 10.11KK70 pKa = 10.66AKK72 pKa = 10.74EE73 pKa = 4.35GIQMVLLLRR82 pKa = 11.84SLQKK86 pKa = 10.62SEE88 pKa = 4.43FATEE92 pKa = 3.87DD93 pKa = 2.73WTLIDD98 pKa = 3.6TSAEE102 pKa = 4.0LLHH105 pKa = 6.36TPPKK109 pKa = 10.58NCFKK113 pKa = 10.85KK114 pKa = 10.39GGYY117 pKa = 6.19TVEE120 pKa = 4.35VYY122 pKa = 10.61FDD124 pKa = 4.13EE125 pKa = 5.66NPDD128 pKa = 3.42NVFPYY133 pKa = 9.85TNWDD137 pKa = 4.19FIYY140 pKa = 11.08YY141 pKa = 9.94QDD143 pKa = 5.04TNEE146 pKa = 4.34KK147 pKa = 6.92WHH149 pKa = 6.06KK150 pKa = 8.62TAGLVDD156 pKa = 3.78YY157 pKa = 10.94DD158 pKa = 3.5GMYY161 pKa = 10.32FQEE164 pKa = 4.53PNGDD168 pKa = 2.88RR169 pKa = 11.84TYY171 pKa = 11.1FQLFEE176 pKa = 5.05KK177 pKa = 10.74DD178 pKa = 2.96ALRR181 pKa = 11.84YY182 pKa = 9.87GSTGRR187 pKa = 11.84WSVQYY192 pKa = 10.6KK193 pKa = 9.66NQTILPSASVSSSTRR208 pKa = 11.84RR209 pKa = 11.84SVDD212 pKa = 3.18EE213 pKa = 4.24PDD215 pKa = 3.5EE216 pKa = 4.37ARR218 pKa = 11.84SGPSTYY224 pKa = 10.23PSTPPQISSRR234 pKa = 11.84RR235 pKa = 11.84SEE237 pKa = 4.03TEE239 pKa = 3.17EE240 pKa = 4.03RR241 pKa = 11.84GLVSSTLQTPPQTSGEE257 pKa = 3.97VRR259 pKa = 11.84LRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84GKK266 pKa = 10.28GEE268 pKa = 3.55RR269 pKa = 11.84TTRR272 pKa = 11.84KK273 pKa = 9.76SPTKK277 pKa = 10.09RR278 pKa = 11.84RR279 pKa = 11.84RR280 pKa = 11.84TDD282 pKa = 3.15EE283 pKa = 3.92TDD285 pKa = 3.36SPGLEE290 pKa = 4.0SVPSPEE296 pKa = 4.55AVGTRR301 pKa = 11.84HH302 pKa = 5.32RR303 pKa = 11.84TVPSTGLSRR312 pKa = 11.84LRR314 pKa = 11.84RR315 pKa = 11.84LQIEE319 pKa = 4.16ARR321 pKa = 11.84DD322 pKa = 3.76PPAIIVTGPANSLKK336 pKa = 9.9CWRR339 pKa = 11.84WRR341 pKa = 11.84LKK343 pKa = 10.51KK344 pKa = 10.35YY345 pKa = 9.53SRR347 pKa = 11.84YY348 pKa = 7.16YY349 pKa = 11.49ARR351 pKa = 11.84MSTVWSWAGEE361 pKa = 4.25VCPKK365 pKa = 10.26ASKK368 pKa = 10.84NRR370 pKa = 11.84MLIAFTDD377 pKa = 3.56EE378 pKa = 4.15TQRR381 pKa = 11.84SVFLKK386 pKa = 10.6LVSLPKK392 pKa = 9.39HH393 pKa = 4.47TTYY396 pKa = 11.56GLAQLGILL404 pKa = 4.22
MM1 pKa = 7.0NQADD5 pKa = 3.68LTARR9 pKa = 11.84FDD11 pKa = 3.97ALQEE15 pKa = 4.17TLMTLYY21 pKa = 10.29EE22 pKa = 4.17RR23 pKa = 11.84NARR26 pKa = 11.84DD27 pKa = 3.6LEE29 pKa = 4.26SQILHH34 pKa = 6.42WEE36 pKa = 4.49TVRR39 pKa = 11.84KK40 pKa = 9.93QYY42 pKa = 9.59VTMYY46 pKa = 9.06YY47 pKa = 10.43ARR49 pKa = 11.84KK50 pKa = 9.55EE51 pKa = 4.26GFRR54 pKa = 11.84SLGMQPLPALTVSEE68 pKa = 4.51YY69 pKa = 10.11KK70 pKa = 10.66AKK72 pKa = 10.74EE73 pKa = 4.35GIQMVLLLRR82 pKa = 11.84SLQKK86 pKa = 10.62SEE88 pKa = 4.43FATEE92 pKa = 3.87DD93 pKa = 2.73WTLIDD98 pKa = 3.6TSAEE102 pKa = 4.0LLHH105 pKa = 6.36TPPKK109 pKa = 10.58NCFKK113 pKa = 10.85KK114 pKa = 10.39GGYY117 pKa = 6.19TVEE120 pKa = 4.35VYY122 pKa = 10.61FDD124 pKa = 4.13EE125 pKa = 5.66NPDD128 pKa = 3.42NVFPYY133 pKa = 9.85TNWDD137 pKa = 4.19FIYY140 pKa = 11.08YY141 pKa = 9.94QDD143 pKa = 5.04TNEE146 pKa = 4.34KK147 pKa = 6.92WHH149 pKa = 6.06KK150 pKa = 8.62TAGLVDD156 pKa = 3.78YY157 pKa = 10.94DD158 pKa = 3.5GMYY161 pKa = 10.32FQEE164 pKa = 4.53PNGDD168 pKa = 2.88RR169 pKa = 11.84TYY171 pKa = 11.1FQLFEE176 pKa = 5.05KK177 pKa = 10.74DD178 pKa = 2.96ALRR181 pKa = 11.84YY182 pKa = 9.87GSTGRR187 pKa = 11.84WSVQYY192 pKa = 10.6KK193 pKa = 9.66NQTILPSASVSSSTRR208 pKa = 11.84RR209 pKa = 11.84SVDD212 pKa = 3.18EE213 pKa = 4.24PDD215 pKa = 3.5EE216 pKa = 4.37ARR218 pKa = 11.84SGPSTYY224 pKa = 10.23PSTPPQISSRR234 pKa = 11.84RR235 pKa = 11.84SEE237 pKa = 4.03TEE239 pKa = 3.17EE240 pKa = 4.03RR241 pKa = 11.84GLVSSTLQTPPQTSGEE257 pKa = 3.97VRR259 pKa = 11.84LRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84GKK266 pKa = 10.28GEE268 pKa = 3.55RR269 pKa = 11.84TTRR272 pKa = 11.84KK273 pKa = 9.76SPTKK277 pKa = 10.09RR278 pKa = 11.84RR279 pKa = 11.84RR280 pKa = 11.84TDD282 pKa = 3.15EE283 pKa = 3.92TDD285 pKa = 3.36SPGLEE290 pKa = 4.0SVPSPEE296 pKa = 4.55AVGTRR301 pKa = 11.84HH302 pKa = 5.32RR303 pKa = 11.84TVPSTGLSRR312 pKa = 11.84LRR314 pKa = 11.84RR315 pKa = 11.84LQIEE319 pKa = 4.16ARR321 pKa = 11.84DD322 pKa = 3.76PPAIIVTGPANSLKK336 pKa = 9.9CWRR339 pKa = 11.84WRR341 pKa = 11.84LKK343 pKa = 10.51KK344 pKa = 10.35YY345 pKa = 9.53SRR347 pKa = 11.84YY348 pKa = 7.16YY349 pKa = 11.49ARR351 pKa = 11.84MSTVWSWAGEE361 pKa = 4.25VCPKK365 pKa = 10.26ASKK368 pKa = 10.84NRR370 pKa = 11.84MLIAFTDD377 pKa = 3.56EE378 pKa = 4.15TQRR381 pKa = 11.84SVFLKK386 pKa = 10.6LVSLPKK392 pKa = 9.39HH393 pKa = 4.47TTYY396 pKa = 11.56GLAQLGILL404 pKa = 4.22
Molecular weight: 46.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2424 |
97 |
603 |
346.3 |
39.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.992 ± 0.394 | 2.228 ± 0.804 |
6.436 ± 0.512 | 6.436 ± 0.256 |
4.579 ± 0.446 | 5.817 ± 0.775 |
1.733 ± 0.307 | 4.909 ± 0.688 |
5.652 ± 0.813 | 9.323 ± 0.673 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.51 | 4.497 ± 0.658 |
6.353 ± 1.047 | 4.084 ± 0.408 |
5.487 ± 0.869 | 7.88 ± 0.428 |
6.518 ± 0.897 | 5.817 ± 0.37 |
1.238 ± 0.314 | 4.125 ± 0.533 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
