
Mycobacterium phage Poenanya
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Cheoctovirus; unclassified Cheoctovirus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 107 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
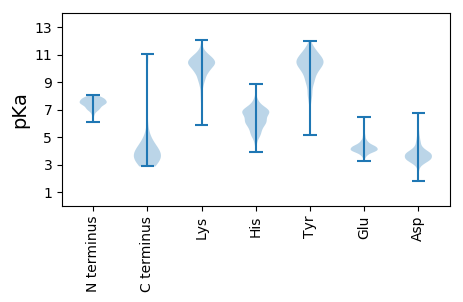
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A411AWY8|A0A411AWY8_9CAUD Lysin B OS=Mycobacterium phage Poenanya OX=2500793 GN=31 PE=4 SV=1
MM1 pKa = 7.38NATEE5 pKa = 5.72DD6 pKa = 3.71GLEE9 pKa = 4.22PLGEE13 pKa = 4.45APDD16 pKa = 4.23LGAPRR21 pKa = 11.84YY22 pKa = 9.14PMSAEE27 pKa = 4.13VTFHH31 pKa = 7.62RR32 pKa = 11.84ARR34 pKa = 11.84CTKK37 pKa = 10.5CGDD40 pKa = 3.08IEE42 pKa = 4.31TDD44 pKa = 3.37YY45 pKa = 11.76GDD47 pKa = 4.22FSAYY51 pKa = 10.11SDD53 pKa = 3.39PGGAINAVLAAADD66 pKa = 3.49WFGRR70 pKa = 11.84SAPTGEE76 pKa = 5.08LIDD79 pKa = 4.08FGGGRR84 pKa = 11.84MGQRR88 pKa = 11.84YY89 pKa = 7.81QLVEE93 pKa = 4.45LLCPDD98 pKa = 3.6CRR100 pKa = 11.84RR101 pKa = 11.84CEE103 pKa = 3.99VCGTAKK109 pKa = 10.44AYY111 pKa = 9.85PIDD114 pKa = 3.85DD115 pKa = 4.0HH116 pKa = 6.93LVCEE120 pKa = 4.29DD121 pKa = 4.12HH122 pKa = 7.29EE123 pKa = 4.31DD124 pKa = 3.94HH125 pKa = 6.79EE126 pKa = 5.25FEE128 pKa = 4.96AAPP131 pKa = 3.95
MM1 pKa = 7.38NATEE5 pKa = 5.72DD6 pKa = 3.71GLEE9 pKa = 4.22PLGEE13 pKa = 4.45APDD16 pKa = 4.23LGAPRR21 pKa = 11.84YY22 pKa = 9.14PMSAEE27 pKa = 4.13VTFHH31 pKa = 7.62RR32 pKa = 11.84ARR34 pKa = 11.84CTKK37 pKa = 10.5CGDD40 pKa = 3.08IEE42 pKa = 4.31TDD44 pKa = 3.37YY45 pKa = 11.76GDD47 pKa = 4.22FSAYY51 pKa = 10.11SDD53 pKa = 3.39PGGAINAVLAAADD66 pKa = 3.49WFGRR70 pKa = 11.84SAPTGEE76 pKa = 5.08LIDD79 pKa = 4.08FGGGRR84 pKa = 11.84MGQRR88 pKa = 11.84YY89 pKa = 7.81QLVEE93 pKa = 4.45LLCPDD98 pKa = 3.6CRR100 pKa = 11.84RR101 pKa = 11.84CEE103 pKa = 3.99VCGTAKK109 pKa = 10.44AYY111 pKa = 9.85PIDD114 pKa = 3.85DD115 pKa = 4.0HH116 pKa = 6.93LVCEE120 pKa = 4.29DD121 pKa = 4.12HH122 pKa = 7.29EE123 pKa = 4.31DD124 pKa = 3.94HH125 pKa = 6.79EE126 pKa = 5.25FEE128 pKa = 4.96AAPP131 pKa = 3.95
Molecular weight: 14.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A411AX56|A0A411AX56_9CAUD Uncharacterized protein OS=Mycobacterium phage Poenanya OX=2500793 GN=91 PE=4 SV=1
MM1 pKa = 6.68RR2 pKa = 11.84TAYY5 pKa = 9.46KK6 pKa = 10.35VRR8 pKa = 11.84AYY10 pKa = 9.99PDD12 pKa = 3.54AEE14 pKa = 3.94QAALLRR20 pKa = 11.84RR21 pKa = 11.84TFGCVRR27 pKa = 11.84LVWNKK32 pKa = 8.63TLAEE36 pKa = 3.94RR37 pKa = 11.84QQRR40 pKa = 11.84YY41 pKa = 5.17TTEE44 pKa = 4.09QKK46 pKa = 8.79STSYY50 pKa = 11.18KK51 pKa = 8.93EE52 pKa = 3.49TDD54 pKa = 3.15AALSEE59 pKa = 4.27WKK61 pKa = 8.75KK62 pKa = 9.31TEE64 pKa = 3.98DD65 pKa = 3.43LAFLSEE71 pKa = 4.44VSSVPLQQTLRR82 pKa = 11.84HH83 pKa = 4.8QHH85 pKa = 5.1SAFAAFFKK93 pKa = 11.02GLAKK97 pKa = 10.48YY98 pKa = 9.91PRR100 pKa = 11.84FKK102 pKa = 10.64SRR104 pKa = 11.84HH105 pKa = 4.52GRR107 pKa = 11.84QSAHH111 pKa = 4.97FTRR114 pKa = 11.84SAFRR118 pKa = 11.84IKK120 pKa = 10.73DD121 pKa = 3.4GALWLAKK128 pKa = 9.15TATPLRR134 pKa = 11.84IVWTWPGVDD143 pKa = 4.93LAALDD148 pKa = 3.81PTMVIVSRR156 pKa = 11.84EE157 pKa = 3.56PDD159 pKa = 3.33GRR161 pKa = 11.84WFVTFAVDD169 pKa = 4.75QPDD172 pKa = 4.38PQPLPATGEE181 pKa = 4.38SVGVDD186 pKa = 3.21LGIKK190 pKa = 10.29DD191 pKa = 4.22FATLSTGEE199 pKa = 4.36KK200 pKa = 9.27IANPRR205 pKa = 11.84HH206 pKa = 5.1MARR209 pKa = 11.84HH210 pKa = 4.89EE211 pKa = 4.24RR212 pKa = 11.84GLRR215 pKa = 11.84RR216 pKa = 11.84QQRR219 pKa = 11.84RR220 pKa = 11.84LSRR223 pKa = 11.84MKK225 pKa = 10.6NGSKK229 pKa = 10.03NRR231 pKa = 11.84ARR233 pKa = 11.84QRR235 pKa = 11.84VKK237 pKa = 10.29VARR240 pKa = 11.84KK241 pKa = 7.65HH242 pKa = 5.79ARR244 pKa = 11.84VRR246 pKa = 11.84DD247 pKa = 3.57ARR249 pKa = 11.84RR250 pKa = 11.84DD251 pKa = 3.87FLHH254 pKa = 6.02KK255 pKa = 9.64TSTEE259 pKa = 3.7LVRR262 pKa = 11.84RR263 pKa = 11.84FDD265 pKa = 4.03TIAVEE270 pKa = 5.02DD271 pKa = 4.2LAPKK275 pKa = 10.67NMVGNRR281 pKa = 11.84SLAKK285 pKa = 10.31SISEE289 pKa = 4.31CGWGEE294 pKa = 3.82FRR296 pKa = 11.84SMLEE300 pKa = 3.95YY301 pKa = 10.21KK302 pKa = 10.32AKK304 pKa = 9.93KK305 pKa = 8.98AGRR308 pKa = 11.84RR309 pKa = 11.84VAVINRR315 pKa = 11.84WYY317 pKa = 9.65PSSKK321 pKa = 8.31TCSACGHH328 pKa = 6.91LLATLSLGTRR338 pKa = 11.84HH339 pKa = 5.33WTCPDD344 pKa = 3.12CGTRR348 pKa = 11.84HH349 pKa = 6.6DD350 pKa = 4.59RR351 pKa = 11.84DD352 pKa = 3.06INAAKK357 pKa = 10.32NILVAAGLAEE367 pKa = 4.11TQNACGGDD375 pKa = 3.84VRR377 pKa = 11.84PHH379 pKa = 6.14GASHH383 pKa = 6.35RR384 pKa = 11.84QSPVKK389 pKa = 10.34QEE391 pKa = 3.98PSQATARR398 pKa = 11.84IPVLQGGEE406 pKa = 3.87
MM1 pKa = 6.68RR2 pKa = 11.84TAYY5 pKa = 9.46KK6 pKa = 10.35VRR8 pKa = 11.84AYY10 pKa = 9.99PDD12 pKa = 3.54AEE14 pKa = 3.94QAALLRR20 pKa = 11.84RR21 pKa = 11.84TFGCVRR27 pKa = 11.84LVWNKK32 pKa = 8.63TLAEE36 pKa = 3.94RR37 pKa = 11.84QQRR40 pKa = 11.84YY41 pKa = 5.17TTEE44 pKa = 4.09QKK46 pKa = 8.79STSYY50 pKa = 11.18KK51 pKa = 8.93EE52 pKa = 3.49TDD54 pKa = 3.15AALSEE59 pKa = 4.27WKK61 pKa = 8.75KK62 pKa = 9.31TEE64 pKa = 3.98DD65 pKa = 3.43LAFLSEE71 pKa = 4.44VSSVPLQQTLRR82 pKa = 11.84HH83 pKa = 4.8QHH85 pKa = 5.1SAFAAFFKK93 pKa = 11.02GLAKK97 pKa = 10.48YY98 pKa = 9.91PRR100 pKa = 11.84FKK102 pKa = 10.64SRR104 pKa = 11.84HH105 pKa = 4.52GRR107 pKa = 11.84QSAHH111 pKa = 4.97FTRR114 pKa = 11.84SAFRR118 pKa = 11.84IKK120 pKa = 10.73DD121 pKa = 3.4GALWLAKK128 pKa = 9.15TATPLRR134 pKa = 11.84IVWTWPGVDD143 pKa = 4.93LAALDD148 pKa = 3.81PTMVIVSRR156 pKa = 11.84EE157 pKa = 3.56PDD159 pKa = 3.33GRR161 pKa = 11.84WFVTFAVDD169 pKa = 4.75QPDD172 pKa = 4.38PQPLPATGEE181 pKa = 4.38SVGVDD186 pKa = 3.21LGIKK190 pKa = 10.29DD191 pKa = 4.22FATLSTGEE199 pKa = 4.36KK200 pKa = 9.27IANPRR205 pKa = 11.84HH206 pKa = 5.1MARR209 pKa = 11.84HH210 pKa = 4.89EE211 pKa = 4.24RR212 pKa = 11.84GLRR215 pKa = 11.84RR216 pKa = 11.84QQRR219 pKa = 11.84RR220 pKa = 11.84LSRR223 pKa = 11.84MKK225 pKa = 10.6NGSKK229 pKa = 10.03NRR231 pKa = 11.84ARR233 pKa = 11.84QRR235 pKa = 11.84VKK237 pKa = 10.29VARR240 pKa = 11.84KK241 pKa = 7.65HH242 pKa = 5.79ARR244 pKa = 11.84VRR246 pKa = 11.84DD247 pKa = 3.57ARR249 pKa = 11.84RR250 pKa = 11.84DD251 pKa = 3.87FLHH254 pKa = 6.02KK255 pKa = 9.64TSTEE259 pKa = 3.7LVRR262 pKa = 11.84RR263 pKa = 11.84FDD265 pKa = 4.03TIAVEE270 pKa = 5.02DD271 pKa = 4.2LAPKK275 pKa = 10.67NMVGNRR281 pKa = 11.84SLAKK285 pKa = 10.31SISEE289 pKa = 4.31CGWGEE294 pKa = 3.82FRR296 pKa = 11.84SMLEE300 pKa = 3.95YY301 pKa = 10.21KK302 pKa = 10.32AKK304 pKa = 9.93KK305 pKa = 8.98AGRR308 pKa = 11.84RR309 pKa = 11.84VAVINRR315 pKa = 11.84WYY317 pKa = 9.65PSSKK321 pKa = 8.31TCSACGHH328 pKa = 6.91LLATLSLGTRR338 pKa = 11.84HH339 pKa = 5.33WTCPDD344 pKa = 3.12CGTRR348 pKa = 11.84HH349 pKa = 6.6DD350 pKa = 4.59RR351 pKa = 11.84DD352 pKa = 3.06INAAKK357 pKa = 10.32NILVAAGLAEE367 pKa = 4.11TQNACGGDD375 pKa = 3.84VRR377 pKa = 11.84PHH379 pKa = 6.14GASHH383 pKa = 6.35RR384 pKa = 11.84QSPVKK389 pKa = 10.34QEE391 pKa = 3.98PSQATARR398 pKa = 11.84IPVLQGGEE406 pKa = 3.87
Molecular weight: 45.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
18568 |
30 |
1226 |
173.5 |
19.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.281 ± 0.428 | 1.217 ± 0.168 |
6.614 ± 0.236 | 5.886 ± 0.302 |
3.054 ± 0.189 | 8.945 ± 0.593 |
2.235 ± 0.171 | 4.346 ± 0.205 |
3.431 ± 0.205 | 7.281 ± 0.222 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.283 ± 0.115 | 3.388 ± 0.175 |
5.967 ± 0.19 | 3.414 ± 0.201 |
6.915 ± 0.45 | 5.897 ± 0.286 |
6.544 ± 0.266 | 7.184 ± 0.266 |
2.429 ± 0.146 | 2.687 ± 0.147 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
