
Spodoptera frugiperda rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betapaprhavirus; Frugiperda betapaprhavirus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
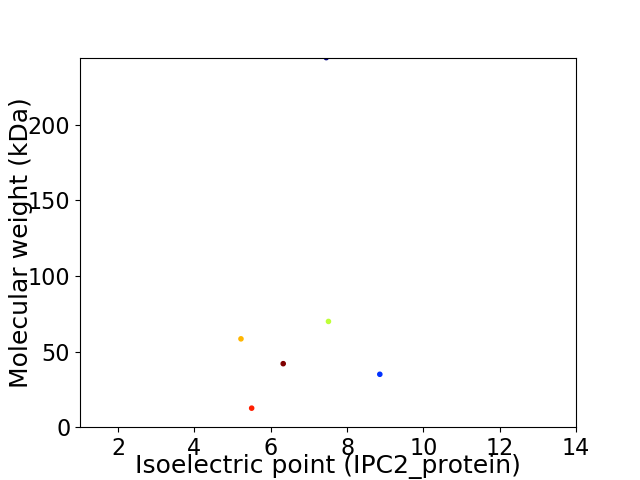
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
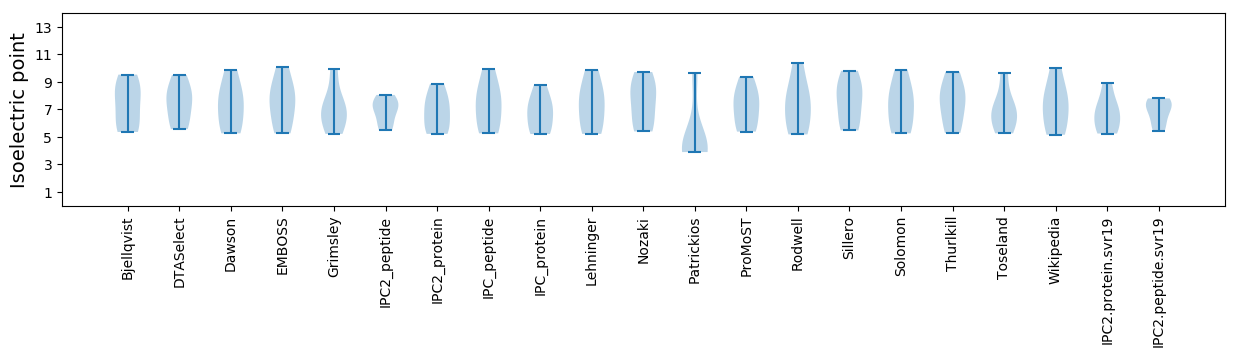
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X2KYE6|X2KYE6_9RHAB Uncharacterized protein OS=Spodoptera frugiperda rhabdovirus OX=1481139 PE=4 SV=1
MM1 pKa = 7.01TQGTMKK7 pKa = 10.35PVWEE11 pKa = 4.04EE12 pKa = 3.76LGTGEE17 pKa = 4.96TEE19 pKa = 4.07FQGTVDD25 pKa = 3.0IPGRR29 pKa = 11.84SLKK32 pKa = 10.25PEE34 pKa = 3.87KK35 pKa = 9.96TDD37 pKa = 3.12WSVDD41 pKa = 3.2TCRR44 pKa = 11.84EE45 pKa = 3.23ISLNLKK51 pKa = 10.35LPGEE55 pKa = 4.61IWQLAHH61 pKa = 6.75QEE63 pKa = 4.4TIFNRR68 pKa = 11.84FLTFYY73 pKa = 10.09ATGYY77 pKa = 9.71VPNTHH82 pKa = 6.43TATEE86 pKa = 4.42IVLSMASLIFKK97 pKa = 10.25DD98 pKa = 3.65KK99 pKa = 11.13AKK101 pKa = 11.04APIDD105 pKa = 4.83LIWDD109 pKa = 3.85DD110 pKa = 4.55SFQASPSEE118 pKa = 4.02EE119 pKa = 4.07CGFSVVGEE127 pKa = 4.3TPLVIGQHH135 pKa = 6.76PDD137 pKa = 3.19DD138 pKa = 5.56DD139 pKa = 5.84DD140 pKa = 3.75YY141 pKa = 11.95TLRR144 pKa = 11.84EE145 pKa = 4.33DD146 pKa = 4.02EE147 pKa = 4.92EE148 pKa = 4.45SAAMNEE154 pKa = 4.12EE155 pKa = 4.17EE156 pKa = 5.28KK157 pKa = 10.73IQAALKK163 pKa = 9.35TLGIQDD169 pKa = 3.94TPVDD173 pKa = 4.26LKK175 pKa = 10.9DD176 pKa = 3.17ASGIVFEE183 pKa = 4.49TKK185 pKa = 9.46EE186 pKa = 4.19DD187 pKa = 3.51RR188 pKa = 11.84EE189 pKa = 3.94QRR191 pKa = 11.84IKK193 pKa = 11.05NEE195 pKa = 3.93KK196 pKa = 9.19ALHH199 pKa = 5.75VEE201 pKa = 4.02DD202 pKa = 6.23DD203 pKa = 3.94INALTQITKK212 pKa = 10.25QFLFEE217 pKa = 4.23YY218 pKa = 9.73STGSLQKK225 pKa = 10.07FVAKK229 pKa = 9.37ATTIFIDD236 pKa = 3.61NNATNGFTRR245 pKa = 11.84LHH247 pKa = 5.53LHH249 pKa = 7.77AIRR252 pKa = 11.84VMNFIALTMLRR263 pKa = 11.84KK264 pKa = 8.3VTKK267 pKa = 10.66SNAQMINAFLKK278 pKa = 9.84EE279 pKa = 3.9QYY281 pKa = 9.71KK282 pKa = 10.83RR283 pKa = 11.84NIASLIPGALSSDD296 pKa = 4.31FAPPSKK302 pKa = 10.86SCIDD306 pKa = 3.43KK307 pKa = 9.33LTAISKK313 pKa = 10.11NDD315 pKa = 3.75PAVSSFFAKK324 pKa = 9.71VVMLNMEE331 pKa = 4.31EE332 pKa = 4.31EE333 pKa = 4.14RR334 pKa = 11.84RR335 pKa = 11.84NPSLVACLGASLLTHH350 pKa = 5.76TTWNGMGILHH360 pKa = 6.71VIFEE364 pKa = 4.72VCLFHH369 pKa = 7.16QISWKK374 pKa = 10.38RR375 pKa = 11.84LVTEE379 pKa = 4.6SLTSLTKK386 pKa = 9.91MSWGEE391 pKa = 3.72VSQFLIKK398 pKa = 10.57YY399 pKa = 6.17QAKK402 pKa = 9.88GNPDD406 pKa = 3.3PTVAWARR413 pKa = 11.84IIDD416 pKa = 3.74DD417 pKa = 4.15SYY419 pKa = 11.8FMRR422 pKa = 11.84LTIVNHH428 pKa = 5.63PTLAALLVEE437 pKa = 4.3SLIRR441 pKa = 11.84SQKK444 pKa = 10.92DD445 pKa = 3.27DD446 pKa = 4.72GILNANWAIQHH457 pKa = 6.93RR458 pKa = 11.84DD459 pKa = 3.18TINYY463 pKa = 8.66YY464 pKa = 10.39RR465 pKa = 11.84DD466 pKa = 3.34AAKK469 pKa = 10.85LLTDD473 pKa = 3.79KK474 pKa = 10.65LTGQTATVQALTNEE488 pKa = 4.17AADD491 pKa = 4.38LVRR494 pKa = 11.84TMNAGPSRR502 pKa = 11.84YY503 pKa = 9.04HH504 pKa = 6.42PRR506 pKa = 11.84PSTLIPMVDD515 pKa = 4.65LNPEE519 pKa = 4.09DD520 pKa = 3.63LL521 pKa = 4.71
MM1 pKa = 7.01TQGTMKK7 pKa = 10.35PVWEE11 pKa = 4.04EE12 pKa = 3.76LGTGEE17 pKa = 4.96TEE19 pKa = 4.07FQGTVDD25 pKa = 3.0IPGRR29 pKa = 11.84SLKK32 pKa = 10.25PEE34 pKa = 3.87KK35 pKa = 9.96TDD37 pKa = 3.12WSVDD41 pKa = 3.2TCRR44 pKa = 11.84EE45 pKa = 3.23ISLNLKK51 pKa = 10.35LPGEE55 pKa = 4.61IWQLAHH61 pKa = 6.75QEE63 pKa = 4.4TIFNRR68 pKa = 11.84FLTFYY73 pKa = 10.09ATGYY77 pKa = 9.71VPNTHH82 pKa = 6.43TATEE86 pKa = 4.42IVLSMASLIFKK97 pKa = 10.25DD98 pKa = 3.65KK99 pKa = 11.13AKK101 pKa = 11.04APIDD105 pKa = 4.83LIWDD109 pKa = 3.85DD110 pKa = 4.55SFQASPSEE118 pKa = 4.02EE119 pKa = 4.07CGFSVVGEE127 pKa = 4.3TPLVIGQHH135 pKa = 6.76PDD137 pKa = 3.19DD138 pKa = 5.56DD139 pKa = 5.84DD140 pKa = 3.75YY141 pKa = 11.95TLRR144 pKa = 11.84EE145 pKa = 4.33DD146 pKa = 4.02EE147 pKa = 4.92EE148 pKa = 4.45SAAMNEE154 pKa = 4.12EE155 pKa = 4.17EE156 pKa = 5.28KK157 pKa = 10.73IQAALKK163 pKa = 9.35TLGIQDD169 pKa = 3.94TPVDD173 pKa = 4.26LKK175 pKa = 10.9DD176 pKa = 3.17ASGIVFEE183 pKa = 4.49TKK185 pKa = 9.46EE186 pKa = 4.19DD187 pKa = 3.51RR188 pKa = 11.84EE189 pKa = 3.94QRR191 pKa = 11.84IKK193 pKa = 11.05NEE195 pKa = 3.93KK196 pKa = 9.19ALHH199 pKa = 5.75VEE201 pKa = 4.02DD202 pKa = 6.23DD203 pKa = 3.94INALTQITKK212 pKa = 10.25QFLFEE217 pKa = 4.23YY218 pKa = 9.73STGSLQKK225 pKa = 10.07FVAKK229 pKa = 9.37ATTIFIDD236 pKa = 3.61NNATNGFTRR245 pKa = 11.84LHH247 pKa = 5.53LHH249 pKa = 7.77AIRR252 pKa = 11.84VMNFIALTMLRR263 pKa = 11.84KK264 pKa = 8.3VTKK267 pKa = 10.66SNAQMINAFLKK278 pKa = 9.84EE279 pKa = 3.9QYY281 pKa = 9.71KK282 pKa = 10.83RR283 pKa = 11.84NIASLIPGALSSDD296 pKa = 4.31FAPPSKK302 pKa = 10.86SCIDD306 pKa = 3.43KK307 pKa = 9.33LTAISKK313 pKa = 10.11NDD315 pKa = 3.75PAVSSFFAKK324 pKa = 9.71VVMLNMEE331 pKa = 4.31EE332 pKa = 4.31EE333 pKa = 4.14RR334 pKa = 11.84RR335 pKa = 11.84NPSLVACLGASLLTHH350 pKa = 5.76TTWNGMGILHH360 pKa = 6.71VIFEE364 pKa = 4.72VCLFHH369 pKa = 7.16QISWKK374 pKa = 10.38RR375 pKa = 11.84LVTEE379 pKa = 4.6SLTSLTKK386 pKa = 9.91MSWGEE391 pKa = 3.72VSQFLIKK398 pKa = 10.57YY399 pKa = 6.17QAKK402 pKa = 9.88GNPDD406 pKa = 3.3PTVAWARR413 pKa = 11.84IIDD416 pKa = 3.74DD417 pKa = 4.15SYY419 pKa = 11.8FMRR422 pKa = 11.84LTIVNHH428 pKa = 5.63PTLAALLVEE437 pKa = 4.3SLIRR441 pKa = 11.84SQKK444 pKa = 10.92DD445 pKa = 3.27DD446 pKa = 4.72GILNANWAIQHH457 pKa = 6.93RR458 pKa = 11.84DD459 pKa = 3.18TINYY463 pKa = 8.66YY464 pKa = 10.39RR465 pKa = 11.84DD466 pKa = 3.34AAKK469 pKa = 10.85LLTDD473 pKa = 3.79KK474 pKa = 10.65LTGQTATVQALTNEE488 pKa = 4.17AADD491 pKa = 4.38LVRR494 pKa = 11.84TMNAGPSRR502 pKa = 11.84YY503 pKa = 9.04HH504 pKa = 6.42PRR506 pKa = 11.84PSTLIPMVDD515 pKa = 4.65LNPEE519 pKa = 4.09DD520 pKa = 3.63LL521 pKa = 4.71
Molecular weight: 58.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X2L5J8|X2L5J8_9RHAB P OS=Spodoptera frugiperda rhabdovirus OX=1481139 PE=4 SV=1
MM1 pKa = 7.43SALEE5 pKa = 4.23RR6 pKa = 11.84IARR9 pKa = 11.84SLSLKK14 pKa = 9.99KK15 pKa = 10.44LNPRR19 pKa = 11.84RR20 pKa = 11.84TPKK23 pKa = 8.86TQPIPEE29 pKa = 4.5KK30 pKa = 9.55ATVYY34 pKa = 10.45HH35 pKa = 7.05PFMLSYY41 pKa = 10.97DD42 pKa = 3.87LNLAIEE48 pKa = 4.47GKK50 pKa = 9.42IHH52 pKa = 6.75ISAITIIVNALSLAWAIEE70 pKa = 4.1LFHH73 pKa = 7.5SDD75 pKa = 4.12SSWSGCLEE83 pKa = 4.09YY84 pKa = 10.28FWKK87 pKa = 10.57SIKK90 pKa = 10.64DD91 pKa = 3.7NILASINPRR100 pKa = 11.84VDD102 pKa = 3.48PNGTCHH108 pKa = 6.2MMTSIITFLGFSDD121 pKa = 4.9GSCINSEE128 pKa = 4.12AEE130 pKa = 3.84PRR132 pKa = 11.84QLTGSRR138 pKa = 11.84SWEE141 pKa = 3.63IMSPNQNLIVITLGFKK157 pKa = 8.95ITLKK161 pKa = 10.11TFAQHH166 pKa = 5.18QRR168 pKa = 11.84YY169 pKa = 8.97SLRR172 pKa = 11.84DD173 pKa = 3.4HH174 pKa = 6.93GFHH177 pKa = 6.83KK178 pKa = 11.08LEE180 pKa = 3.99MLNEE184 pKa = 4.15KK185 pKa = 8.95EE186 pKa = 4.65KK187 pKa = 11.37KK188 pKa = 7.62MLNYY192 pKa = 9.91MGVKK196 pKa = 8.96QLKK199 pKa = 8.38PQYY202 pKa = 7.38THH204 pKa = 6.55EE205 pKa = 4.23KK206 pKa = 8.95TFEE209 pKa = 3.81KK210 pKa = 10.8LILKK214 pKa = 10.22NKK216 pKa = 8.39GPKK219 pKa = 9.36GSRR222 pKa = 11.84VRR224 pKa = 11.84AILHH228 pKa = 4.88SQSRR232 pKa = 11.84DD233 pKa = 2.95MWSPTAPSPPPTYY246 pKa = 10.53EE247 pKa = 5.03DD248 pKa = 3.88GSSDD252 pKa = 3.88EE253 pKa = 4.23WDD255 pKa = 3.42QQQLHH260 pKa = 6.22SLNHH264 pKa = 5.73LHH266 pKa = 6.15TPSVPLRR273 pKa = 11.84APRR276 pKa = 11.84TSPPQQLSPKK286 pKa = 8.23PTSTTQPLPQLTQPNKK302 pKa = 9.45PQEE305 pKa = 3.96LSKK308 pKa = 11.34
MM1 pKa = 7.43SALEE5 pKa = 4.23RR6 pKa = 11.84IARR9 pKa = 11.84SLSLKK14 pKa = 9.99KK15 pKa = 10.44LNPRR19 pKa = 11.84RR20 pKa = 11.84TPKK23 pKa = 8.86TQPIPEE29 pKa = 4.5KK30 pKa = 9.55ATVYY34 pKa = 10.45HH35 pKa = 7.05PFMLSYY41 pKa = 10.97DD42 pKa = 3.87LNLAIEE48 pKa = 4.47GKK50 pKa = 9.42IHH52 pKa = 6.75ISAITIIVNALSLAWAIEE70 pKa = 4.1LFHH73 pKa = 7.5SDD75 pKa = 4.12SSWSGCLEE83 pKa = 4.09YY84 pKa = 10.28FWKK87 pKa = 10.57SIKK90 pKa = 10.64DD91 pKa = 3.7NILASINPRR100 pKa = 11.84VDD102 pKa = 3.48PNGTCHH108 pKa = 6.2MMTSIITFLGFSDD121 pKa = 4.9GSCINSEE128 pKa = 4.12AEE130 pKa = 3.84PRR132 pKa = 11.84QLTGSRR138 pKa = 11.84SWEE141 pKa = 3.63IMSPNQNLIVITLGFKK157 pKa = 8.95ITLKK161 pKa = 10.11TFAQHH166 pKa = 5.18QRR168 pKa = 11.84YY169 pKa = 8.97SLRR172 pKa = 11.84DD173 pKa = 3.4HH174 pKa = 6.93GFHH177 pKa = 6.83KK178 pKa = 11.08LEE180 pKa = 3.99MLNEE184 pKa = 4.15KK185 pKa = 8.95EE186 pKa = 4.65KK187 pKa = 11.37KK188 pKa = 7.62MLNYY192 pKa = 9.91MGVKK196 pKa = 8.96QLKK199 pKa = 8.38PQYY202 pKa = 7.38THH204 pKa = 6.55EE205 pKa = 4.23KK206 pKa = 8.95TFEE209 pKa = 3.81KK210 pKa = 10.8LILKK214 pKa = 10.22NKK216 pKa = 8.39GPKK219 pKa = 9.36GSRR222 pKa = 11.84VRR224 pKa = 11.84AILHH228 pKa = 4.88SQSRR232 pKa = 11.84DD233 pKa = 2.95MWSPTAPSPPPTYY246 pKa = 10.53EE247 pKa = 5.03DD248 pKa = 3.88GSSDD252 pKa = 3.88EE253 pKa = 4.23WDD255 pKa = 3.42QQQLHH260 pKa = 6.22SLNHH264 pKa = 5.73LHH266 pKa = 6.15TPSVPLRR273 pKa = 11.84APRR276 pKa = 11.84TSPPQQLSPKK286 pKa = 8.23PTSTTQPLPQLTQPNKK302 pKa = 9.45PQEE305 pKa = 3.96LSKK308 pKa = 11.34
Molecular weight: 34.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4069 |
109 |
2140 |
678.2 |
77.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.596 ± 1.018 | 1.425 ± 0.195 |
5.087 ± 0.481 | 5.235 ± 0.322 |
4.375 ± 0.603 | 4.792 ± 0.391 |
2.875 ± 0.175 | 6.906 ± 0.586 |
5.628 ± 0.274 | 11.969 ± 1.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.284 | 4.989 ± 0.194 |
5.825 ± 0.453 | 4.178 ± 0.296 |
4.841 ± 0.32 | 8.675 ± 0.72 |
6.267 ± 0.753 | 5.48 ± 0.493 |
1.597 ± 0.254 | 3.244 ± 0.402 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
