
Duck circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 7.99
Get precalculated fractions of proteins
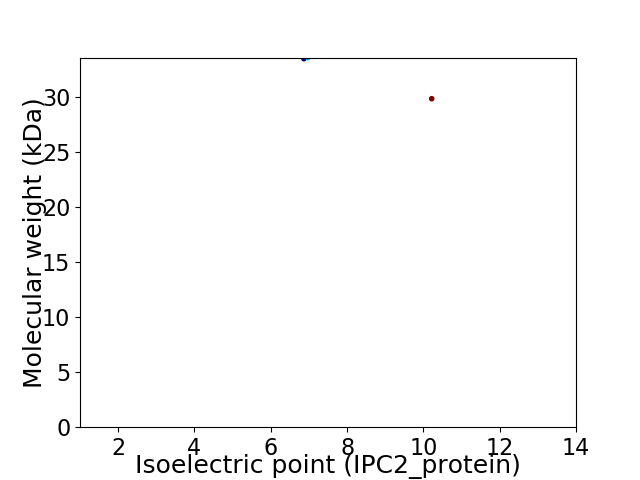
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
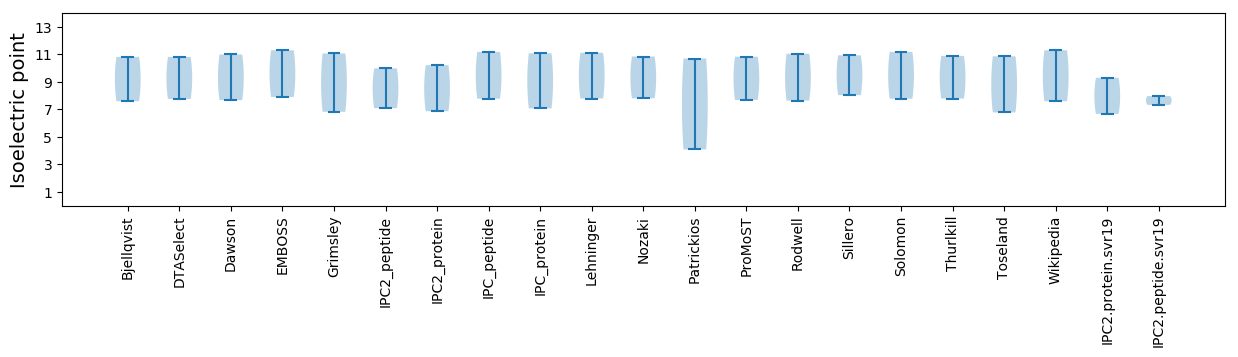
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I2CZK4|I2CZK4_9CIRC Capsid protein OS=Duck circovirus OX=324685 GN=Cap PE=3 SV=1
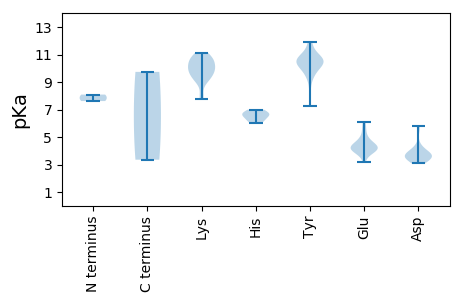
MM1 pKa = 7.62AKK3 pKa = 10.03SGNYY7 pKa = 9.34SYY9 pKa = 11.03KK10 pKa = 10.22RR11 pKa = 11.84WVFTINNPTFEE22 pKa = 5.38DD23 pKa = 4.37YY24 pKa = 11.38VHH26 pKa = 6.99VLEE29 pKa = 6.12FCTLDD34 pKa = 3.2NCKK37 pKa = 9.85FAIVGEE43 pKa = 4.22EE44 pKa = 3.75KK45 pKa = 10.66GANGTPHH52 pKa = 6.68LQGFLNLRR60 pKa = 11.84SNARR64 pKa = 11.84AAALEE69 pKa = 3.96EE70 pKa = 4.42SLGGRR75 pKa = 11.84AWLSRR80 pKa = 11.84ARR82 pKa = 11.84GSDD85 pKa = 3.31EE86 pKa = 5.71DD87 pKa = 4.0NEE89 pKa = 4.41EE90 pKa = 4.14YY91 pKa = 10.26CAKK94 pKa = 10.09EE95 pKa = 3.72STYY98 pKa = 10.94LRR100 pKa = 11.84VGEE103 pKa = 4.2PVSKK107 pKa = 10.61GRR109 pKa = 11.84SSDD112 pKa = 3.5LAEE115 pKa = 4.15ATSAVMAGVPLTEE128 pKa = 4.41VARR131 pKa = 11.84KK132 pKa = 9.21FPTTYY137 pKa = 10.62VIFGRR142 pKa = 11.84GLEE145 pKa = 4.13RR146 pKa = 11.84LRR148 pKa = 11.84HH149 pKa = 6.05LIVEE153 pKa = 4.56TQRR156 pKa = 11.84DD157 pKa = 3.66WKK159 pKa = 9.91TEE161 pKa = 4.0VIVLIGPPGTGKK173 pKa = 10.04SRR175 pKa = 11.84YY176 pKa = 9.4AFEE179 pKa = 4.95FPAEE183 pKa = 3.93NKK185 pKa = 9.86YY186 pKa = 10.59YY187 pKa = 10.65KK188 pKa = 10.16PRR190 pKa = 11.84GKK192 pKa = 8.91WWDD195 pKa = 3.9GYY197 pKa = 10.85SGNDD201 pKa = 3.3VVVMDD206 pKa = 5.83DD207 pKa = 3.98FYY209 pKa = 11.92GWLPYY214 pKa = 10.72DD215 pKa = 4.0DD216 pKa = 5.54LLRR219 pKa = 11.84ITDD222 pKa = 4.33RR223 pKa = 11.84YY224 pKa = 8.36PLRR227 pKa = 11.84VEE229 pKa = 4.85FKK231 pKa = 10.75GGMTQFVAKK240 pKa = 9.46TLIITSNRR248 pKa = 11.84EE249 pKa = 3.17PRR251 pKa = 11.84DD252 pKa = 3.33WYY254 pKa = 10.83KK255 pKa = 11.13SEE257 pKa = 4.41FDD259 pKa = 3.61LSALYY264 pKa = 10.34RR265 pKa = 11.84RR266 pKa = 11.84INKK269 pKa = 9.29YY270 pKa = 10.12LVYY273 pKa = 10.73NIDD276 pKa = 3.46KK277 pKa = 10.73YY278 pKa = 11.39EE279 pKa = 4.14PAQACTLPFPINYY292 pKa = 9.74
MM1 pKa = 7.62AKK3 pKa = 10.03SGNYY7 pKa = 9.34SYY9 pKa = 11.03KK10 pKa = 10.22RR11 pKa = 11.84WVFTINNPTFEE22 pKa = 5.38DD23 pKa = 4.37YY24 pKa = 11.38VHH26 pKa = 6.99VLEE29 pKa = 6.12FCTLDD34 pKa = 3.2NCKK37 pKa = 9.85FAIVGEE43 pKa = 4.22EE44 pKa = 3.75KK45 pKa = 10.66GANGTPHH52 pKa = 6.68LQGFLNLRR60 pKa = 11.84SNARR64 pKa = 11.84AAALEE69 pKa = 3.96EE70 pKa = 4.42SLGGRR75 pKa = 11.84AWLSRR80 pKa = 11.84ARR82 pKa = 11.84GSDD85 pKa = 3.31EE86 pKa = 5.71DD87 pKa = 4.0NEE89 pKa = 4.41EE90 pKa = 4.14YY91 pKa = 10.26CAKK94 pKa = 10.09EE95 pKa = 3.72STYY98 pKa = 10.94LRR100 pKa = 11.84VGEE103 pKa = 4.2PVSKK107 pKa = 10.61GRR109 pKa = 11.84SSDD112 pKa = 3.5LAEE115 pKa = 4.15ATSAVMAGVPLTEE128 pKa = 4.41VARR131 pKa = 11.84KK132 pKa = 9.21FPTTYY137 pKa = 10.62VIFGRR142 pKa = 11.84GLEE145 pKa = 4.13RR146 pKa = 11.84LRR148 pKa = 11.84HH149 pKa = 6.05LIVEE153 pKa = 4.56TQRR156 pKa = 11.84DD157 pKa = 3.66WKK159 pKa = 9.91TEE161 pKa = 4.0VIVLIGPPGTGKK173 pKa = 10.04SRR175 pKa = 11.84YY176 pKa = 9.4AFEE179 pKa = 4.95FPAEE183 pKa = 3.93NKK185 pKa = 9.86YY186 pKa = 10.59YY187 pKa = 10.65KK188 pKa = 10.16PRR190 pKa = 11.84GKK192 pKa = 8.91WWDD195 pKa = 3.9GYY197 pKa = 10.85SGNDD201 pKa = 3.3VVVMDD206 pKa = 5.83DD207 pKa = 3.98FYY209 pKa = 11.92GWLPYY214 pKa = 10.72DD215 pKa = 4.0DD216 pKa = 5.54LLRR219 pKa = 11.84ITDD222 pKa = 4.33RR223 pKa = 11.84YY224 pKa = 8.36PLRR227 pKa = 11.84VEE229 pKa = 4.85FKK231 pKa = 10.75GGMTQFVAKK240 pKa = 9.46TLIITSNRR248 pKa = 11.84EE249 pKa = 3.17PRR251 pKa = 11.84DD252 pKa = 3.33WYY254 pKa = 10.83KK255 pKa = 11.13SEE257 pKa = 4.41FDD259 pKa = 3.61LSALYY264 pKa = 10.34RR265 pKa = 11.84RR266 pKa = 11.84INKK269 pKa = 9.29YY270 pKa = 10.12LVYY273 pKa = 10.73NIDD276 pKa = 3.46KK277 pKa = 10.73YY278 pKa = 11.39EE279 pKa = 4.14PAQACTLPFPINYY292 pKa = 9.74
Molecular weight: 33.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I2CZK4|I2CZK4_9CIRC Capsid protein OS=Duck circovirus OX=324685 GN=Cap PE=3 SV=1
MM1 pKa = 8.09RR2 pKa = 11.84GRR4 pKa = 11.84TYY6 pKa = 10.46RR7 pKa = 11.84RR8 pKa = 11.84AYY10 pKa = 9.23RR11 pKa = 11.84GRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.89RR16 pKa = 11.84RR17 pKa = 11.84GLRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84LRR29 pKa = 11.84IARR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84FSVVTYY42 pKa = 10.07KK43 pKa = 9.3VTRR46 pKa = 11.84NTVFGFFGSQTGPTAAGKK64 pKa = 8.0WQSLSLEE71 pKa = 4.75DD72 pKa = 3.59GAQYY76 pKa = 10.5TDD78 pKa = 3.7PPARR82 pKa = 11.84GNNICGLNMRR92 pKa = 11.84WAMFGDD98 pKa = 3.86TNSYY102 pKa = 7.24MTGSTPFYY110 pKa = 9.79HH111 pKa = 6.53YY112 pKa = 10.3PYY114 pKa = 9.79DD115 pKa = 3.64YY116 pKa = 11.93YY117 pKa = 10.47MIKK120 pKa = 10.47GVAITLRR127 pKa = 11.84PAYY130 pKa = 10.29NIYY133 pKa = 10.31QKK135 pKa = 11.11SKK137 pKa = 7.74TQGSTVIDD145 pKa = 3.45KK146 pKa = 10.75DD147 pKa = 3.68GQIVKK152 pKa = 9.55TSTTGWSIDD161 pKa = 3.79PYY163 pKa = 10.79GSTSSRR169 pKa = 11.84RR170 pKa = 11.84TWDD173 pKa = 3.07PSRR176 pKa = 11.84VHH178 pKa = 6.3RR179 pKa = 11.84RR180 pKa = 11.84YY181 pKa = 10.13FIPKK185 pKa = 9.62PIIQRR190 pKa = 11.84AGEE193 pKa = 4.15GTKK196 pKa = 10.74YY197 pKa = 9.78STFFLGGRR205 pKa = 11.84NFTWINCAQDD215 pKa = 3.56QVVHH219 pKa = 6.74YY220 pKa = 10.14GMGMSLRR227 pKa = 11.84KK228 pKa = 9.19PDD230 pKa = 3.16NTTGVNAQYY239 pKa = 10.62DD240 pKa = 3.7IEE242 pKa = 4.33AQFTFYY248 pKa = 10.69IKK250 pKa = 10.53FGQFTGFF257 pKa = 3.35
MM1 pKa = 8.09RR2 pKa = 11.84GRR4 pKa = 11.84TYY6 pKa = 10.46RR7 pKa = 11.84RR8 pKa = 11.84AYY10 pKa = 9.23RR11 pKa = 11.84GRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.89RR16 pKa = 11.84RR17 pKa = 11.84GLRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84LRR29 pKa = 11.84IARR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84FSVVTYY42 pKa = 10.07KK43 pKa = 9.3VTRR46 pKa = 11.84NTVFGFFGSQTGPTAAGKK64 pKa = 8.0WQSLSLEE71 pKa = 4.75DD72 pKa = 3.59GAQYY76 pKa = 10.5TDD78 pKa = 3.7PPARR82 pKa = 11.84GNNICGLNMRR92 pKa = 11.84WAMFGDD98 pKa = 3.86TNSYY102 pKa = 7.24MTGSTPFYY110 pKa = 9.79HH111 pKa = 6.53YY112 pKa = 10.3PYY114 pKa = 9.79DD115 pKa = 3.64YY116 pKa = 11.93YY117 pKa = 10.47MIKK120 pKa = 10.47GVAITLRR127 pKa = 11.84PAYY130 pKa = 10.29NIYY133 pKa = 10.31QKK135 pKa = 11.11SKK137 pKa = 7.74TQGSTVIDD145 pKa = 3.45KK146 pKa = 10.75DD147 pKa = 3.68GQIVKK152 pKa = 9.55TSTTGWSIDD161 pKa = 3.79PYY163 pKa = 10.79GSTSSRR169 pKa = 11.84RR170 pKa = 11.84TWDD173 pKa = 3.07PSRR176 pKa = 11.84VHH178 pKa = 6.3RR179 pKa = 11.84RR180 pKa = 11.84YY181 pKa = 10.13FIPKK185 pKa = 9.62PIIQRR190 pKa = 11.84AGEE193 pKa = 4.15GTKK196 pKa = 10.74YY197 pKa = 9.78STFFLGGRR205 pKa = 11.84NFTWINCAQDD215 pKa = 3.56QVVHH219 pKa = 6.74YY220 pKa = 10.14GMGMSLRR227 pKa = 11.84KK228 pKa = 9.19PDD230 pKa = 3.16NTTGVNAQYY239 pKa = 10.62DD240 pKa = 3.7IEE242 pKa = 4.33AQFTFYY248 pKa = 10.69IKK250 pKa = 10.53FGQFTGFF257 pKa = 3.35
Molecular weight: 29.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
549 |
257 |
292 |
274.5 |
31.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.193 ± 0.762 | 1.093 ± 0.211 |
4.918 ± 0.428 | 4.554 ± 2.275 |
5.464 ± 0.511 | 8.743 ± 0.923 |
1.093 ± 0.05 | 4.736 ± 0.478 |
5.282 ± 0.412 | 5.829 ± 1.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.004 ± 0.484 | 4.372 ± 0.323 |
4.918 ± 0.167 | 2.914 ± 1.179 |
10.018 ± 1.896 | 5.829 ± 0.267 |
7.65 ± 1.395 | 5.464 ± 0.795 |
2.186 ± 0.161 | 6.74 ± 0.178 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
