
Sanxia water strider virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Lispiviridae; Arlivirus; Gerrid arlivirus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
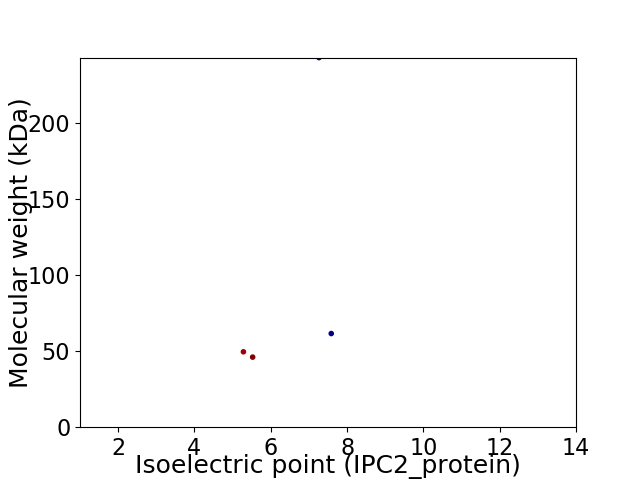
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXD7|A0A0B5KXD7_9MONO ORF2 OS=Sanxia water strider virus 4 OX=1608063 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.83ANIDD5 pKa = 4.43DD6 pKa = 4.14NLEE9 pKa = 3.98TTFRR13 pKa = 11.84RR14 pKa = 11.84SIKK17 pKa = 9.75KK18 pKa = 9.53NKK20 pKa = 8.77RR21 pKa = 11.84DD22 pKa = 3.43RR23 pKa = 11.84VLTSDD28 pKa = 3.77CSVLINLIGSLHH40 pKa = 5.34TLIEE44 pKa = 4.22NTGGKK49 pKa = 9.56VNPALKK55 pKa = 10.53KK56 pKa = 7.26MTSNAPHH63 pKa = 6.83DD64 pKa = 4.71NIVEE68 pKa = 4.17EE69 pKa = 4.76EE70 pKa = 4.35GSGMKK75 pKa = 8.69EE76 pKa = 4.0TIEE79 pKa = 4.23GFQSQEE85 pKa = 3.79FEE87 pKa = 4.65LNDD90 pKa = 4.32LLVEE94 pKa = 4.76DD95 pKa = 4.68PSKK98 pKa = 10.66LTVEE102 pKa = 4.62MAQVMRR108 pKa = 11.84EE109 pKa = 4.28SISEE113 pKa = 3.91HH114 pKa = 6.61RR115 pKa = 11.84DD116 pKa = 3.02YY117 pKa = 11.41DD118 pKa = 3.88SEE120 pKa = 4.57VIQGEE125 pKa = 4.2AQDD128 pKa = 4.54LEE130 pKa = 4.84VPAVDD135 pKa = 5.21MRR137 pKa = 11.84DD138 pKa = 3.42EE139 pKa = 4.66DD140 pKa = 5.35LSLLDD145 pKa = 5.74DD146 pKa = 4.3VIEE149 pKa = 4.91PYY151 pKa = 10.74EE152 pKa = 4.73HH153 pKa = 6.25MWQMYY158 pKa = 6.48THH160 pKa = 6.93VLTLIEE166 pKa = 4.21RR167 pKa = 11.84EE168 pKa = 4.13EE169 pKa = 4.45IKK171 pKa = 10.93NAWDD175 pKa = 3.67TIYY178 pKa = 11.11NFYY181 pKa = 10.59PEE183 pKa = 4.79EE184 pKa = 4.5SPLTRR189 pKa = 11.84FNTLMLVKK197 pKa = 10.46LQDD200 pKa = 3.25HH201 pKa = 5.2VHH203 pKa = 5.7RR204 pKa = 11.84EE205 pKa = 3.83KK206 pKa = 11.04RR207 pKa = 11.84LVEE210 pKa = 3.98QVDD213 pKa = 3.71RR214 pKa = 11.84KK215 pKa = 10.52LDD217 pKa = 3.66SLSTKK222 pKa = 9.1VTVDD226 pKa = 2.74TDD228 pKa = 3.98NIIKK232 pKa = 9.68EE233 pKa = 4.03IKK235 pKa = 9.72KK236 pKa = 9.61IQIKK240 pKa = 6.34TTSTPIPAIAVSSPSTSKK258 pKa = 11.07NINLPKK264 pKa = 10.43AGTGTPTISGDD275 pKa = 3.81DD276 pKa = 3.74FLNKK280 pKa = 10.19LKK282 pKa = 10.42EE283 pKa = 3.86LRR285 pKa = 11.84KK286 pKa = 10.48NKK288 pKa = 7.78TAPVKK293 pKa = 10.04RR294 pKa = 11.84DD295 pKa = 3.17VVIIPKK301 pKa = 8.45EE302 pKa = 3.97TLKK305 pKa = 10.88FNPIILGPEE314 pKa = 4.15VQSKK318 pKa = 9.61PVVRR322 pKa = 11.84FTKK325 pKa = 10.42DD326 pKa = 3.1VSRR329 pKa = 11.84VSAAVIMEE337 pKa = 4.47EE338 pKa = 4.45KK339 pKa = 10.79SKK341 pKa = 9.92TDD343 pKa = 3.36KK344 pKa = 11.32YY345 pKa = 9.29WDD347 pKa = 3.61KK348 pKa = 11.08LISSGFLLDD357 pKa = 3.77RR358 pKa = 11.84EE359 pKa = 4.6KK360 pKa = 11.31VDD362 pKa = 4.26ALYY365 pKa = 11.29NSLKK369 pKa = 10.66LNKK372 pKa = 9.72EE373 pKa = 4.18LFNGLLDD380 pKa = 4.28PNNLDD385 pKa = 3.65RR386 pKa = 11.84SHH388 pKa = 6.38HH389 pKa = 5.86QGMIDD394 pKa = 4.01YY395 pKa = 9.4IWKK398 pKa = 9.78EE399 pKa = 3.7LGDD402 pKa = 3.64VGKK405 pKa = 10.63QSDD408 pKa = 4.68FIDD411 pKa = 4.75LMNHH415 pKa = 5.28MKK417 pKa = 8.97KK418 pKa = 6.75TTYY421 pKa = 9.3FVAIIRR427 pKa = 11.84YY428 pKa = 7.44WNLLEE433 pKa = 4.17
MM1 pKa = 7.83ANIDD5 pKa = 4.43DD6 pKa = 4.14NLEE9 pKa = 3.98TTFRR13 pKa = 11.84RR14 pKa = 11.84SIKK17 pKa = 9.75KK18 pKa = 9.53NKK20 pKa = 8.77RR21 pKa = 11.84DD22 pKa = 3.43RR23 pKa = 11.84VLTSDD28 pKa = 3.77CSVLINLIGSLHH40 pKa = 5.34TLIEE44 pKa = 4.22NTGGKK49 pKa = 9.56VNPALKK55 pKa = 10.53KK56 pKa = 7.26MTSNAPHH63 pKa = 6.83DD64 pKa = 4.71NIVEE68 pKa = 4.17EE69 pKa = 4.76EE70 pKa = 4.35GSGMKK75 pKa = 8.69EE76 pKa = 4.0TIEE79 pKa = 4.23GFQSQEE85 pKa = 3.79FEE87 pKa = 4.65LNDD90 pKa = 4.32LLVEE94 pKa = 4.76DD95 pKa = 4.68PSKK98 pKa = 10.66LTVEE102 pKa = 4.62MAQVMRR108 pKa = 11.84EE109 pKa = 4.28SISEE113 pKa = 3.91HH114 pKa = 6.61RR115 pKa = 11.84DD116 pKa = 3.02YY117 pKa = 11.41DD118 pKa = 3.88SEE120 pKa = 4.57VIQGEE125 pKa = 4.2AQDD128 pKa = 4.54LEE130 pKa = 4.84VPAVDD135 pKa = 5.21MRR137 pKa = 11.84DD138 pKa = 3.42EE139 pKa = 4.66DD140 pKa = 5.35LSLLDD145 pKa = 5.74DD146 pKa = 4.3VIEE149 pKa = 4.91PYY151 pKa = 10.74EE152 pKa = 4.73HH153 pKa = 6.25MWQMYY158 pKa = 6.48THH160 pKa = 6.93VLTLIEE166 pKa = 4.21RR167 pKa = 11.84EE168 pKa = 4.13EE169 pKa = 4.45IKK171 pKa = 10.93NAWDD175 pKa = 3.67TIYY178 pKa = 11.11NFYY181 pKa = 10.59PEE183 pKa = 4.79EE184 pKa = 4.5SPLTRR189 pKa = 11.84FNTLMLVKK197 pKa = 10.46LQDD200 pKa = 3.25HH201 pKa = 5.2VHH203 pKa = 5.7RR204 pKa = 11.84EE205 pKa = 3.83KK206 pKa = 11.04RR207 pKa = 11.84LVEE210 pKa = 3.98QVDD213 pKa = 3.71RR214 pKa = 11.84KK215 pKa = 10.52LDD217 pKa = 3.66SLSTKK222 pKa = 9.1VTVDD226 pKa = 2.74TDD228 pKa = 3.98NIIKK232 pKa = 9.68EE233 pKa = 4.03IKK235 pKa = 9.72KK236 pKa = 9.61IQIKK240 pKa = 6.34TTSTPIPAIAVSSPSTSKK258 pKa = 11.07NINLPKK264 pKa = 10.43AGTGTPTISGDD275 pKa = 3.81DD276 pKa = 3.74FLNKK280 pKa = 10.19LKK282 pKa = 10.42EE283 pKa = 3.86LRR285 pKa = 11.84KK286 pKa = 10.48NKK288 pKa = 7.78TAPVKK293 pKa = 10.04RR294 pKa = 11.84DD295 pKa = 3.17VVIIPKK301 pKa = 8.45EE302 pKa = 3.97TLKK305 pKa = 10.88FNPIILGPEE314 pKa = 4.15VQSKK318 pKa = 9.61PVVRR322 pKa = 11.84FTKK325 pKa = 10.42DD326 pKa = 3.1VSRR329 pKa = 11.84VSAAVIMEE337 pKa = 4.47EE338 pKa = 4.45KK339 pKa = 10.79SKK341 pKa = 9.92TDD343 pKa = 3.36KK344 pKa = 11.32YY345 pKa = 9.29WDD347 pKa = 3.61KK348 pKa = 11.08LISSGFLLDD357 pKa = 3.77RR358 pKa = 11.84EE359 pKa = 4.6KK360 pKa = 11.31VDD362 pKa = 4.26ALYY365 pKa = 11.29NSLKK369 pKa = 10.66LNKK372 pKa = 9.72EE373 pKa = 4.18LFNGLLDD380 pKa = 4.28PNNLDD385 pKa = 3.65RR386 pKa = 11.84SHH388 pKa = 6.38HH389 pKa = 5.86QGMIDD394 pKa = 4.01YY395 pKa = 9.4IWKK398 pKa = 9.78EE399 pKa = 3.7LGDD402 pKa = 3.64VGKK405 pKa = 10.63QSDD408 pKa = 4.68FIDD411 pKa = 4.75LMNHH415 pKa = 5.28MKK417 pKa = 8.97KK418 pKa = 6.75TTYY421 pKa = 9.3FVAIIRR427 pKa = 11.84YY428 pKa = 7.44WNLLEE433 pKa = 4.17
Molecular weight: 49.62 kDa
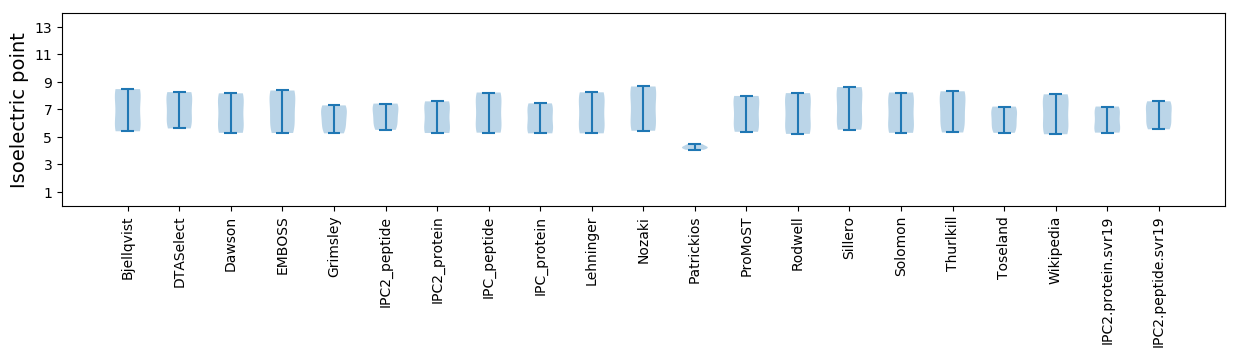
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KK30|A0A0B5KK30_9MONO RNA-directed RNA polymerase L OS=Sanxia water strider virus 4 OX=1608063 GN=L PE=3 SV=1
MM1 pKa = 7.33VSNHH5 pKa = 5.3SWKK8 pKa = 10.41CVIIILVNFTTMHH21 pKa = 6.98GDD23 pKa = 4.68LIFPDD28 pKa = 3.51IGYY31 pKa = 8.28MCEE34 pKa = 3.79PTRR37 pKa = 11.84FMMFSTGSEE46 pKa = 4.12TFHH49 pKa = 7.82IGLKK53 pKa = 10.14LPNIRR58 pKa = 11.84HH59 pKa = 6.26KK60 pKa = 11.1GADD63 pKa = 3.92CIKK66 pKa = 10.12EE67 pKa = 4.13SPEE70 pKa = 3.7VSNLLEE76 pKa = 4.15EE77 pKa = 5.92LYY79 pKa = 10.88DD80 pKa = 4.02LPQKK84 pKa = 10.38VIPNMNFLKK93 pKa = 10.47EE94 pKa = 3.73ISPRR98 pKa = 11.84SNFTRR103 pKa = 11.84DD104 pKa = 3.03KK105 pKa = 11.02RR106 pKa = 11.84FLTAIIGVVSLLSLGTSLYY125 pKa = 11.14GLGKK129 pKa = 10.11QYY131 pKa = 11.14SFEE134 pKa = 4.35SDD136 pKa = 2.63IDD138 pKa = 3.89DD139 pKa = 4.56KK140 pKa = 11.31IRR142 pKa = 11.84RR143 pKa = 11.84QDD145 pKa = 3.47LALHH149 pKa = 6.07EE150 pKa = 5.15LMIKK154 pKa = 10.21HH155 pKa = 5.68NQLLTVLDD163 pKa = 4.06EE164 pKa = 4.64QSNEE168 pKa = 3.81LKK170 pKa = 10.99NLGSSVDD177 pKa = 3.44MLHH180 pKa = 7.47RR181 pKa = 11.84IIEE184 pKa = 4.75NITCIQDD191 pKa = 3.36EE192 pKa = 4.97KK193 pKa = 10.59IQQLEE198 pKa = 4.59TITYY202 pKa = 8.32IQFFILKK209 pKa = 9.82LEE211 pKa = 4.27SFVYY215 pKa = 10.26AALKK219 pKa = 10.63GDD221 pKa = 4.03LHH223 pKa = 7.02PDD225 pKa = 3.45LVTLSMIRR233 pKa = 11.84TLIKK237 pKa = 10.14QRR239 pKa = 11.84SSLRR243 pKa = 11.84DD244 pKa = 3.34TLLDD248 pKa = 3.72KK249 pKa = 11.2DD250 pKa = 3.91PSVLYY255 pKa = 10.58NVGKK259 pKa = 10.36VIPLSYY265 pKa = 10.48NHH267 pKa = 6.34AHH269 pKa = 6.66RR270 pKa = 11.84VITLMIIMPIIRR282 pKa = 11.84LDD284 pKa = 4.04QISHH288 pKa = 7.18IYY290 pKa = 9.96DD291 pKa = 3.55CSNIGFKK298 pKa = 10.76KK299 pKa = 10.59NNTLFKK305 pKa = 9.98YY306 pKa = 7.72TLPEE310 pKa = 4.02NIITVKK316 pKa = 10.48VDD318 pKa = 3.43GKK320 pKa = 10.77QVMTSVNLQYY330 pKa = 10.96CHH332 pKa = 6.44TSGIIVMCNVNAIPTTNYY350 pKa = 10.13QCLDD354 pKa = 3.66DD355 pKa = 5.11LNNNVSSNCSPKK367 pKa = 10.73LLDD370 pKa = 3.75NLLPVAINTQQHH382 pKa = 4.15IMLRR386 pKa = 11.84GFSRR390 pKa = 11.84YY391 pKa = 8.93SVHH394 pKa = 5.62TQSTIPRR401 pKa = 11.84YY402 pKa = 10.01GSYY405 pKa = 10.47KK406 pKa = 10.26INLNPDD412 pKa = 3.16QNTVIAIDD420 pKa = 3.39KK421 pKa = 8.78GKK423 pKa = 10.44KK424 pKa = 8.91YY425 pKa = 10.94VVGGDD430 pKa = 2.7IFTNNVVKK438 pKa = 10.72ANYY441 pKa = 8.75DD442 pKa = 3.13IPTNLSIKK450 pKa = 9.87TMVKK454 pKa = 10.4NISLNNKK461 pKa = 9.08EE462 pKa = 4.59LDD464 pKa = 4.09NISKK468 pKa = 10.06EE469 pKa = 4.04IQKK472 pKa = 10.3LRR474 pKa = 11.84IEE476 pKa = 5.09NISDD480 pKa = 3.3WDD482 pKa = 3.67VSDD485 pKa = 3.88PSTYY489 pKa = 10.73HH490 pKa = 7.0FITIYY495 pKa = 10.35TLLLVSIAALLAISWCYY512 pKa = 10.68LRR514 pKa = 11.84GHH516 pKa = 7.98LIRR519 pKa = 11.84KK520 pKa = 8.01NKK522 pKa = 9.44SKK524 pKa = 10.63KK525 pKa = 8.71FKK527 pKa = 10.31KK528 pKa = 9.99IVMQHH533 pKa = 5.65SNNEE537 pKa = 3.96DD538 pKa = 3.03SDD540 pKa = 3.84
MM1 pKa = 7.33VSNHH5 pKa = 5.3SWKK8 pKa = 10.41CVIIILVNFTTMHH21 pKa = 6.98GDD23 pKa = 4.68LIFPDD28 pKa = 3.51IGYY31 pKa = 8.28MCEE34 pKa = 3.79PTRR37 pKa = 11.84FMMFSTGSEE46 pKa = 4.12TFHH49 pKa = 7.82IGLKK53 pKa = 10.14LPNIRR58 pKa = 11.84HH59 pKa = 6.26KK60 pKa = 11.1GADD63 pKa = 3.92CIKK66 pKa = 10.12EE67 pKa = 4.13SPEE70 pKa = 3.7VSNLLEE76 pKa = 4.15EE77 pKa = 5.92LYY79 pKa = 10.88DD80 pKa = 4.02LPQKK84 pKa = 10.38VIPNMNFLKK93 pKa = 10.47EE94 pKa = 3.73ISPRR98 pKa = 11.84SNFTRR103 pKa = 11.84DD104 pKa = 3.03KK105 pKa = 11.02RR106 pKa = 11.84FLTAIIGVVSLLSLGTSLYY125 pKa = 11.14GLGKK129 pKa = 10.11QYY131 pKa = 11.14SFEE134 pKa = 4.35SDD136 pKa = 2.63IDD138 pKa = 3.89DD139 pKa = 4.56KK140 pKa = 11.31IRR142 pKa = 11.84RR143 pKa = 11.84QDD145 pKa = 3.47LALHH149 pKa = 6.07EE150 pKa = 5.15LMIKK154 pKa = 10.21HH155 pKa = 5.68NQLLTVLDD163 pKa = 4.06EE164 pKa = 4.64QSNEE168 pKa = 3.81LKK170 pKa = 10.99NLGSSVDD177 pKa = 3.44MLHH180 pKa = 7.47RR181 pKa = 11.84IIEE184 pKa = 4.75NITCIQDD191 pKa = 3.36EE192 pKa = 4.97KK193 pKa = 10.59IQQLEE198 pKa = 4.59TITYY202 pKa = 8.32IQFFILKK209 pKa = 9.82LEE211 pKa = 4.27SFVYY215 pKa = 10.26AALKK219 pKa = 10.63GDD221 pKa = 4.03LHH223 pKa = 7.02PDD225 pKa = 3.45LVTLSMIRR233 pKa = 11.84TLIKK237 pKa = 10.14QRR239 pKa = 11.84SSLRR243 pKa = 11.84DD244 pKa = 3.34TLLDD248 pKa = 3.72KK249 pKa = 11.2DD250 pKa = 3.91PSVLYY255 pKa = 10.58NVGKK259 pKa = 10.36VIPLSYY265 pKa = 10.48NHH267 pKa = 6.34AHH269 pKa = 6.66RR270 pKa = 11.84VITLMIIMPIIRR282 pKa = 11.84LDD284 pKa = 4.04QISHH288 pKa = 7.18IYY290 pKa = 9.96DD291 pKa = 3.55CSNIGFKK298 pKa = 10.76KK299 pKa = 10.59NNTLFKK305 pKa = 9.98YY306 pKa = 7.72TLPEE310 pKa = 4.02NIITVKK316 pKa = 10.48VDD318 pKa = 3.43GKK320 pKa = 10.77QVMTSVNLQYY330 pKa = 10.96CHH332 pKa = 6.44TSGIIVMCNVNAIPTTNYY350 pKa = 10.13QCLDD354 pKa = 3.66DD355 pKa = 5.11LNNNVSSNCSPKK367 pKa = 10.73LLDD370 pKa = 3.75NLLPVAINTQQHH382 pKa = 4.15IMLRR386 pKa = 11.84GFSRR390 pKa = 11.84YY391 pKa = 8.93SVHH394 pKa = 5.62TQSTIPRR401 pKa = 11.84YY402 pKa = 10.01GSYY405 pKa = 10.47KK406 pKa = 10.26INLNPDD412 pKa = 3.16QNTVIAIDD420 pKa = 3.39KK421 pKa = 8.78GKK423 pKa = 10.44KK424 pKa = 8.91YY425 pKa = 10.94VVGGDD430 pKa = 2.7IFTNNVVKK438 pKa = 10.72ANYY441 pKa = 8.75DD442 pKa = 3.13IPTNLSIKK450 pKa = 9.87TMVKK454 pKa = 10.4NISLNNKK461 pKa = 9.08EE462 pKa = 4.59LDD464 pKa = 4.09NISKK468 pKa = 10.06EE469 pKa = 4.04IQKK472 pKa = 10.3LRR474 pKa = 11.84IEE476 pKa = 5.09NISDD480 pKa = 3.3WDD482 pKa = 3.67VSDD485 pKa = 3.88PSTYY489 pKa = 10.73HH490 pKa = 7.0FITIYY495 pKa = 10.35TLLLVSIAALLAISWCYY512 pKa = 10.68LRR514 pKa = 11.84GHH516 pKa = 7.98LIRR519 pKa = 11.84KK520 pKa = 8.01NKK522 pKa = 9.44SKK524 pKa = 10.63KK525 pKa = 8.71FKK527 pKa = 10.31KK528 pKa = 9.99IVMQHH533 pKa = 5.65SNNEE537 pKa = 3.96DD538 pKa = 3.03SDD540 pKa = 3.84
Molecular weight: 61.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3489 |
408 |
2108 |
872.3 |
100.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.783 ± 0.519 | 1.748 ± 0.327 |
5.818 ± 0.696 | 6.048 ± 0.726 |
4.385 ± 0.499 | 4.328 ± 0.199 |
2.522 ± 0.15 | 8.885 ± 0.512 |
7.223 ± 0.511 | 11.235 ± 0.327 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.752 ± 0.085 | 5.99 ± 0.464 |
3.239 ± 0.258 | 3.124 ± 0.214 |
4.471 ± 0.253 | 7.509 ± 0.374 |
6.22 ± 0.455 | 5.876 ± 0.34 |
1.146 ± 0.22 | 3.697 ± 0.413 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
