
Burkholderia glumae (strain BGR1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Burkholderia; Burkholderia glumae
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
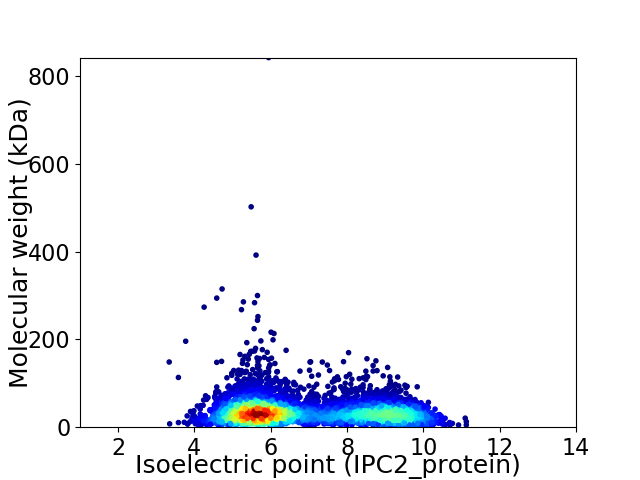
Virtual 2D-PAGE plot for 5660 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5AN22|C5AN22_BURGB Transcriptional regulator/antitoxin MazE OS=Burkholderia glumae (strain BGR1) OX=626418 GN=bglu_1p0480 PE=4 SV=1
MM1 pKa = 6.57TTLTVGGYY9 pKa = 10.0SLGNLDD15 pKa = 5.05GISSVAAAAQQYY27 pKa = 10.1NDD29 pKa = 3.51NRR31 pKa = 11.84TTSNLINYY39 pKa = 7.3QAALANAAASATSLIPGEE57 pKa = 4.32GAAFAGNAIAANIKK71 pKa = 10.16NLSQNHH77 pKa = 4.76STMSASDD84 pKa = 3.87FEE86 pKa = 4.68SSAIAIAGALATLVGQGVEE105 pKa = 4.0ISGLAAANINPLVGGGVFTLGEE127 pKa = 4.44FINAVGFVISAAGVVIDD144 pKa = 4.83SGTIAQIATQSLQALGLAPSPSGQYY169 pKa = 10.78SPVTLSSSNFICDD182 pKa = 3.25ATNAATMWDD191 pKa = 3.43IVKK194 pKa = 10.61GDD196 pKa = 3.45NTEE199 pKa = 4.35QINSAPSNNGSGLNISNSIGGDD221 pKa = 3.21IIVAPGSPGSPYY233 pKa = 9.76TFSISGSDD241 pKa = 3.67NNIMGINSTPITGSAVGVNVSGSGNTINAGTGTSVSLSNTNGNFDD286 pKa = 3.83VVNSNGDD293 pKa = 3.19QFNQGGGISLASNTQANVNGNGNGISVANGDD324 pKa = 3.6SMGAYY329 pKa = 10.16GGGNTINAASNSLVVASSTNGAFDD353 pKa = 4.55TINTNGNTFGGTVANGQGTGVWLNQDD379 pKa = 2.45AQANVNGNNNGISVTSGDD397 pKa = 3.39SMGAYY402 pKa = 10.17GGGNTINAASNSLVVASGTNGSYY425 pKa = 9.37DD426 pKa = 3.58TINANGNSFGGTTANGQGTGVWLDD450 pKa = 3.59KK451 pKa = 11.06NVQANVNGNNIGISVTSGDD470 pKa = 3.39SMGAYY475 pKa = 10.17GGGNTINATPNSVVYY490 pKa = 10.5ASGTNGSYY498 pKa = 9.37DD499 pKa = 3.58TINANGNSFGGAAANGQGTGVWLDD523 pKa = 3.59KK524 pKa = 11.06NVQANVNGNNIGISVTSGDD543 pKa = 3.39SMGAYY548 pKa = 10.17GGGNTINATPNSVVYY563 pKa = 10.5ASGTNGSYY571 pKa = 9.37DD572 pKa = 3.58TINANGNSFGGAAANGQGTGVWLDD596 pKa = 3.59KK597 pKa = 11.06NVQANVNGNNIGISVTSGDD616 pKa = 3.39SMGAYY621 pKa = 10.17GGGNTINATPNSVVYY636 pKa = 10.5ASGTNGSYY644 pKa = 9.37DD645 pKa = 3.58TINANGNSFGGTAANGQGTGVWLDD669 pKa = 3.44KK670 pKa = 11.0NVQVNVNGAGNGIALNSGDD689 pKa = 3.79SVGVYY694 pKa = 10.6GGGNVVDD701 pKa = 4.74AAPITTTAIYY711 pKa = 7.72NTNGNAVVVNATGDD725 pKa = 3.48LFGAVTANGQGSGIWLGQNAQATVNGNGNGIGLTAGATLTATGNSDD771 pKa = 3.17IVNAAAGSVVNVSGTGDD788 pKa = 3.89EE789 pKa = 4.5INGSNSIINIAGNNEE804 pKa = 4.35TIWLSGEE811 pKa = 4.41SNVVTVTGQNVTVYY825 pKa = 10.41AQNSTVNFVGSAANNNNNKK844 pKa = 10.4LIGVKK849 pKa = 8.8NTGSGWSSNDD859 pKa = 2.21AWYY862 pKa = 10.59ALTGSSHH869 pKa = 7.59PIGATAPFQLPVFTDD884 pKa = 4.51PSASVPGDD892 pKa = 3.52NVPNATDD899 pKa = 3.81PVWIFIPYY907 pKa = 8.35TYY909 pKa = 10.31SPSGEE914 pKa = 4.6PILSEE919 pKa = 4.46PIIVTAPPDD928 pKa = 3.6VGDD931 pKa = 4.24SDD933 pKa = 5.91PIILNLNGDD942 pKa = 4.04KK943 pKa = 11.1VQTTALEE950 pKa = 4.53GSTTYY955 pKa = 11.04FDD957 pKa = 4.12MEE959 pKa = 4.35NEE961 pKa = 4.26GEE963 pKa = 4.23KK964 pKa = 10.76NQTAWGTTGEE974 pKa = 4.69GYY976 pKa = 10.55LVYY979 pKa = 10.71DD980 pKa = 4.69PEE982 pKa = 5.81DD983 pKa = 3.44VDD985 pKa = 3.66NAAVVTKK992 pKa = 10.3DD993 pKa = 3.17AQLVGGFGALQTMAQQFDD1011 pKa = 4.27GTGHH1015 pKa = 7.03GDD1017 pKa = 3.53LTVADD1022 pKa = 4.66ALWANLKK1029 pKa = 9.88VWVDD1033 pKa = 3.38KK1034 pKa = 10.82TGSGNFQSGQLMSLDD1049 pKa = 3.53QLGITSIDD1057 pKa = 3.54LDD1059 pKa = 4.06GNQTDD1064 pKa = 4.27RR1065 pKa = 11.84NSNGNKK1071 pKa = 9.86IITDD1075 pKa = 3.64SSFTRR1080 pKa = 11.84ADD1082 pKa = 3.36GSRR1085 pKa = 11.84GDD1087 pKa = 3.47IAGVDD1092 pKa = 3.76LMFSRR1097 pKa = 11.84NDD1099 pKa = 3.41TPSLVDD1105 pKa = 3.57AQVHH1109 pKa = 5.17NLISAMSLAGAAPGGQTSTGVADD1132 pKa = 4.16QNNLAALALPSHH1144 pKa = 6.97
MM1 pKa = 6.57TTLTVGGYY9 pKa = 10.0SLGNLDD15 pKa = 5.05GISSVAAAAQQYY27 pKa = 10.1NDD29 pKa = 3.51NRR31 pKa = 11.84TTSNLINYY39 pKa = 7.3QAALANAAASATSLIPGEE57 pKa = 4.32GAAFAGNAIAANIKK71 pKa = 10.16NLSQNHH77 pKa = 4.76STMSASDD84 pKa = 3.87FEE86 pKa = 4.68SSAIAIAGALATLVGQGVEE105 pKa = 4.0ISGLAAANINPLVGGGVFTLGEE127 pKa = 4.44FINAVGFVISAAGVVIDD144 pKa = 4.83SGTIAQIATQSLQALGLAPSPSGQYY169 pKa = 10.78SPVTLSSSNFICDD182 pKa = 3.25ATNAATMWDD191 pKa = 3.43IVKK194 pKa = 10.61GDD196 pKa = 3.45NTEE199 pKa = 4.35QINSAPSNNGSGLNISNSIGGDD221 pKa = 3.21IIVAPGSPGSPYY233 pKa = 9.76TFSISGSDD241 pKa = 3.67NNIMGINSTPITGSAVGVNVSGSGNTINAGTGTSVSLSNTNGNFDD286 pKa = 3.83VVNSNGDD293 pKa = 3.19QFNQGGGISLASNTQANVNGNGNGISVANGDD324 pKa = 3.6SMGAYY329 pKa = 10.16GGGNTINAASNSLVVASSTNGAFDD353 pKa = 4.55TINTNGNTFGGTVANGQGTGVWLNQDD379 pKa = 2.45AQANVNGNNNGISVTSGDD397 pKa = 3.39SMGAYY402 pKa = 10.17GGGNTINAASNSLVVASGTNGSYY425 pKa = 9.37DD426 pKa = 3.58TINANGNSFGGTTANGQGTGVWLDD450 pKa = 3.59KK451 pKa = 11.06NVQANVNGNNIGISVTSGDD470 pKa = 3.39SMGAYY475 pKa = 10.17GGGNTINATPNSVVYY490 pKa = 10.5ASGTNGSYY498 pKa = 9.37DD499 pKa = 3.58TINANGNSFGGAAANGQGTGVWLDD523 pKa = 3.59KK524 pKa = 11.06NVQANVNGNNIGISVTSGDD543 pKa = 3.39SMGAYY548 pKa = 10.17GGGNTINATPNSVVYY563 pKa = 10.5ASGTNGSYY571 pKa = 9.37DD572 pKa = 3.58TINANGNSFGGAAANGQGTGVWLDD596 pKa = 3.59KK597 pKa = 11.06NVQANVNGNNIGISVTSGDD616 pKa = 3.39SMGAYY621 pKa = 10.17GGGNTINATPNSVVYY636 pKa = 10.5ASGTNGSYY644 pKa = 9.37DD645 pKa = 3.58TINANGNSFGGTAANGQGTGVWLDD669 pKa = 3.44KK670 pKa = 11.0NVQVNVNGAGNGIALNSGDD689 pKa = 3.79SVGVYY694 pKa = 10.6GGGNVVDD701 pKa = 4.74AAPITTTAIYY711 pKa = 7.72NTNGNAVVVNATGDD725 pKa = 3.48LFGAVTANGQGSGIWLGQNAQATVNGNGNGIGLTAGATLTATGNSDD771 pKa = 3.17IVNAAAGSVVNVSGTGDD788 pKa = 3.89EE789 pKa = 4.5INGSNSIINIAGNNEE804 pKa = 4.35TIWLSGEE811 pKa = 4.41SNVVTVTGQNVTVYY825 pKa = 10.41AQNSTVNFVGSAANNNNNKK844 pKa = 10.4LIGVKK849 pKa = 8.8NTGSGWSSNDD859 pKa = 2.21AWYY862 pKa = 10.59ALTGSSHH869 pKa = 7.59PIGATAPFQLPVFTDD884 pKa = 4.51PSASVPGDD892 pKa = 3.52NVPNATDD899 pKa = 3.81PVWIFIPYY907 pKa = 8.35TYY909 pKa = 10.31SPSGEE914 pKa = 4.6PILSEE919 pKa = 4.46PIIVTAPPDD928 pKa = 3.6VGDD931 pKa = 4.24SDD933 pKa = 5.91PIILNLNGDD942 pKa = 4.04KK943 pKa = 11.1VQTTALEE950 pKa = 4.53GSTTYY955 pKa = 11.04FDD957 pKa = 4.12MEE959 pKa = 4.35NEE961 pKa = 4.26GEE963 pKa = 4.23KK964 pKa = 10.76NQTAWGTTGEE974 pKa = 4.69GYY976 pKa = 10.55LVYY979 pKa = 10.71DD980 pKa = 4.69PEE982 pKa = 5.81DD983 pKa = 3.44VDD985 pKa = 3.66NAAVVTKK992 pKa = 10.3DD993 pKa = 3.17AQLVGGFGALQTMAQQFDD1011 pKa = 4.27GTGHH1015 pKa = 7.03GDD1017 pKa = 3.53LTVADD1022 pKa = 4.66ALWANLKK1029 pKa = 9.88VWVDD1033 pKa = 3.38KK1034 pKa = 10.82TGSGNFQSGQLMSLDD1049 pKa = 3.53QLGITSIDD1057 pKa = 3.54LDD1059 pKa = 4.06GNQTDD1064 pKa = 4.27RR1065 pKa = 11.84NSNGNKK1071 pKa = 9.86IITDD1075 pKa = 3.64SSFTRR1080 pKa = 11.84ADD1082 pKa = 3.36GSRR1085 pKa = 11.84GDD1087 pKa = 3.47IAGVDD1092 pKa = 3.76LMFSRR1097 pKa = 11.84NDD1099 pKa = 3.41TPSLVDD1105 pKa = 3.57AQVHH1109 pKa = 5.17NLISAMSLAGAAPGGQTSTGVADD1132 pKa = 4.16QNNLAALALPSHH1144 pKa = 6.97
Molecular weight: 113.28 kDa
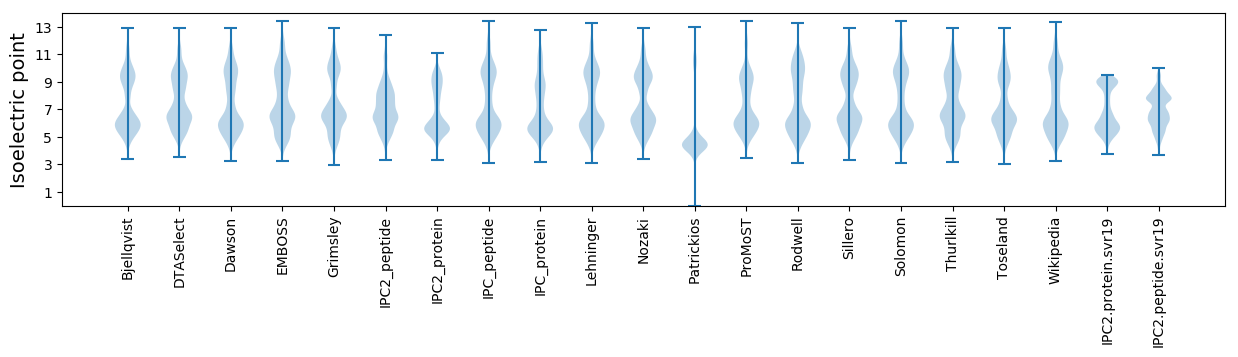
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5AEN1|C5AEN1_BURGB Na+-driven multidrug efflux pump OS=Burkholderia glumae (strain BGR1) OX=626418 GN=bglu_1g11880 PE=4 SV=1
MM1 pKa = 7.98PLRR4 pKa = 11.84LPSRR8 pKa = 11.84PNRR11 pKa = 11.84AARR14 pKa = 11.84SLSRR18 pKa = 11.84LDD20 pKa = 3.38ARR22 pKa = 11.84RR23 pKa = 11.84TARR26 pKa = 11.84TLRR29 pKa = 11.84HH30 pKa = 4.77HH31 pKa = 5.71QARR34 pKa = 11.84ARR36 pKa = 11.84ALATRR41 pKa = 11.84PAAPTSRR48 pKa = 11.84LDD50 pKa = 3.56GLRR53 pKa = 11.84GWLHH57 pKa = 6.28SLLATMRR64 pKa = 11.84AHH66 pKa = 7.31AGARR70 pKa = 11.84RR71 pKa = 11.84LGLPAALLRR80 pKa = 11.84KK81 pKa = 9.28PSSRR85 pKa = 11.84RR86 pKa = 11.84HH87 pKa = 3.91AHH89 pKa = 5.15GPRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84VNRR97 pKa = 11.84PRR99 pKa = 11.84RR100 pKa = 11.84LAASAGWFAFGTRR113 pKa = 3.72
MM1 pKa = 7.98PLRR4 pKa = 11.84LPSRR8 pKa = 11.84PNRR11 pKa = 11.84AARR14 pKa = 11.84SLSRR18 pKa = 11.84LDD20 pKa = 3.38ARR22 pKa = 11.84RR23 pKa = 11.84TARR26 pKa = 11.84TLRR29 pKa = 11.84HH30 pKa = 4.77HH31 pKa = 5.71QARR34 pKa = 11.84ARR36 pKa = 11.84ALATRR41 pKa = 11.84PAAPTSRR48 pKa = 11.84LDD50 pKa = 3.56GLRR53 pKa = 11.84GWLHH57 pKa = 6.28SLLATMRR64 pKa = 11.84AHH66 pKa = 7.31AGARR70 pKa = 11.84RR71 pKa = 11.84LGLPAALLRR80 pKa = 11.84KK81 pKa = 9.28PSSRR85 pKa = 11.84RR86 pKa = 11.84HH87 pKa = 3.91AHH89 pKa = 5.15GPRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84VNRR97 pKa = 11.84PRR99 pKa = 11.84RR100 pKa = 11.84LAASAGWFAFGTRR113 pKa = 3.72
Molecular weight: 12.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1927278 |
25 |
7806 |
340.5 |
36.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.074 ± 0.059 | 0.927 ± 0.01 |
5.499 ± 0.027 | 5.024 ± 0.034 |
3.462 ± 0.02 | 8.422 ± 0.035 |
2.306 ± 0.016 | 4.337 ± 0.024 |
2.626 ± 0.029 | 10.333 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.127 ± 0.014 | 2.522 ± 0.027 |
5.382 ± 0.031 | 3.499 ± 0.024 |
7.652 ± 0.045 | 5.403 ± 0.046 |
5.225 ± 0.036 | 7.428 ± 0.028 |
1.38 ± 0.013 | 2.371 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
