
Citrus leprosis virus N
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Dichorhavirus; Citrus leprosis N dichorhavirus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
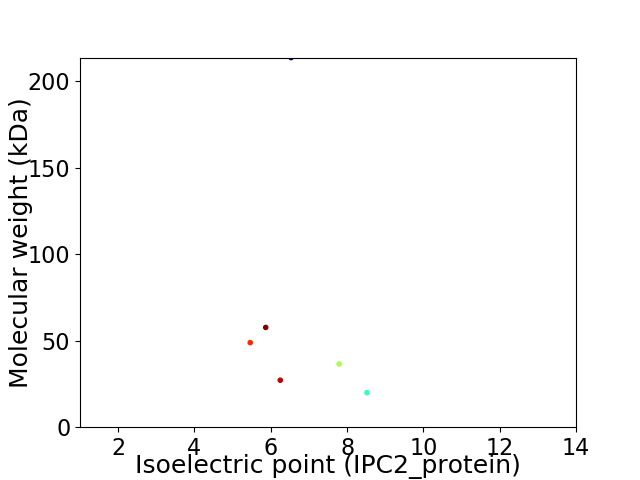
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6AWM3|A0A1W6AWM3_9RHAB Phosphoprotein OS=Citrus leprosis virus N OX=1956177 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.55ASSVMIDD8 pKa = 3.58YY9 pKa = 7.67TTVIPAYY16 pKa = 10.04QATPTEE22 pKa = 4.17NQVASSAPTPKK33 pKa = 10.28QYY35 pKa = 10.28TRR37 pKa = 11.84AAAVAVPIVKK47 pKa = 10.0IPPPTDD53 pKa = 3.11NPQDD57 pKa = 3.3LVTRR61 pKa = 11.84FEE63 pKa = 4.53TLMSTASSILTKK75 pKa = 10.82AEE77 pKa = 3.69LAEE80 pKa = 3.83IMGIGFCITPPGMDD94 pKa = 4.09EE95 pKa = 4.0PAMMMVAVGAVTQGLTMGQIPATYY119 pKa = 10.05DD120 pKa = 3.08SRR122 pKa = 11.84AAAANAMINIDD133 pKa = 3.84PDD135 pKa = 3.63EE136 pKa = 5.17EE137 pKa = 4.29EE138 pKa = 5.29DD139 pKa = 5.34KK140 pKa = 11.47SDD142 pKa = 3.79TPKK145 pKa = 10.88KK146 pKa = 9.61PAEE149 pKa = 3.92EE150 pKa = 4.27SAGSKK155 pKa = 9.86AAAITYY161 pKa = 9.66ICLSLLRR168 pKa = 11.84LCVKK172 pKa = 10.35EE173 pKa = 3.65VDD175 pKa = 3.65TFLKK179 pKa = 10.66GVQQLKK185 pKa = 10.23SSYY188 pKa = 8.97HH189 pKa = 7.18ILMGAHH195 pKa = 6.1SEE197 pKa = 4.35YY198 pKa = 8.35MTSFQYY204 pKa = 10.81SRR206 pKa = 11.84GMCSNIASLFNQCEE220 pKa = 4.07DD221 pKa = 3.25LRR223 pKa = 11.84RR224 pKa = 11.84TLAHH228 pKa = 6.4HH229 pKa = 6.55CAIADD234 pKa = 3.76EE235 pKa = 5.13TYY237 pKa = 8.57HH238 pKa = 5.88QARR241 pKa = 11.84NVHH244 pKa = 5.68GPLRR248 pKa = 11.84FLILQHH254 pKa = 6.3MDD256 pKa = 3.23LTGMIPYY263 pKa = 10.65GMYY266 pKa = 9.83IDD268 pKa = 5.41LKK270 pKa = 10.37MGLPLLSPGLLLTWLHH286 pKa = 6.74DD287 pKa = 3.51AHH289 pKa = 6.96VSNVLALISEE299 pKa = 4.54INTKK303 pKa = 10.38YY304 pKa = 9.55DD305 pKa = 3.21TLTSGDD311 pKa = 3.72RR312 pKa = 11.84FWRR315 pKa = 11.84YY316 pKa = 9.71SRR318 pKa = 11.84GIDD321 pKa = 3.15PGFFLPLQQSKK332 pKa = 10.89CYY334 pKa = 10.39ILIARR339 pKa = 11.84MADD342 pKa = 2.76ILVRR346 pKa = 11.84SGIVSVTQYY355 pKa = 11.13SDD357 pKa = 3.18PRR359 pKa = 11.84NARR362 pKa = 11.84CIADD366 pKa = 3.54KK367 pKa = 8.94TTVKK371 pKa = 10.56AQAEE375 pKa = 4.37IFGKK379 pKa = 9.04EE380 pKa = 3.4FWAAYY385 pKa = 9.79SSLSGSSADD394 pKa = 3.56SGPVAKK400 pKa = 9.72TIASGSRR407 pKa = 11.84TGNITVKK414 pKa = 10.64RR415 pKa = 11.84NLHH418 pKa = 6.0PRR420 pKa = 11.84APPPAPVVNMPPPQSSAPGALDD442 pKa = 3.76EE443 pKa = 5.13MVDD446 pKa = 4.42DD447 pKa = 6.78DD448 pKa = 4.56EE449 pKa = 4.95AQQ451 pKa = 3.07
MM1 pKa = 7.55ASSVMIDD8 pKa = 3.58YY9 pKa = 7.67TTVIPAYY16 pKa = 10.04QATPTEE22 pKa = 4.17NQVASSAPTPKK33 pKa = 10.28QYY35 pKa = 10.28TRR37 pKa = 11.84AAAVAVPIVKK47 pKa = 10.0IPPPTDD53 pKa = 3.11NPQDD57 pKa = 3.3LVTRR61 pKa = 11.84FEE63 pKa = 4.53TLMSTASSILTKK75 pKa = 10.82AEE77 pKa = 3.69LAEE80 pKa = 3.83IMGIGFCITPPGMDD94 pKa = 4.09EE95 pKa = 4.0PAMMMVAVGAVTQGLTMGQIPATYY119 pKa = 10.05DD120 pKa = 3.08SRR122 pKa = 11.84AAAANAMINIDD133 pKa = 3.84PDD135 pKa = 3.63EE136 pKa = 5.17EE137 pKa = 4.29EE138 pKa = 5.29DD139 pKa = 5.34KK140 pKa = 11.47SDD142 pKa = 3.79TPKK145 pKa = 10.88KK146 pKa = 9.61PAEE149 pKa = 3.92EE150 pKa = 4.27SAGSKK155 pKa = 9.86AAAITYY161 pKa = 9.66ICLSLLRR168 pKa = 11.84LCVKK172 pKa = 10.35EE173 pKa = 3.65VDD175 pKa = 3.65TFLKK179 pKa = 10.66GVQQLKK185 pKa = 10.23SSYY188 pKa = 8.97HH189 pKa = 7.18ILMGAHH195 pKa = 6.1SEE197 pKa = 4.35YY198 pKa = 8.35MTSFQYY204 pKa = 10.81SRR206 pKa = 11.84GMCSNIASLFNQCEE220 pKa = 4.07DD221 pKa = 3.25LRR223 pKa = 11.84RR224 pKa = 11.84TLAHH228 pKa = 6.4HH229 pKa = 6.55CAIADD234 pKa = 3.76EE235 pKa = 5.13TYY237 pKa = 8.57HH238 pKa = 5.88QARR241 pKa = 11.84NVHH244 pKa = 5.68GPLRR248 pKa = 11.84FLILQHH254 pKa = 6.3MDD256 pKa = 3.23LTGMIPYY263 pKa = 10.65GMYY266 pKa = 9.83IDD268 pKa = 5.41LKK270 pKa = 10.37MGLPLLSPGLLLTWLHH286 pKa = 6.74DD287 pKa = 3.51AHH289 pKa = 6.96VSNVLALISEE299 pKa = 4.54INTKK303 pKa = 10.38YY304 pKa = 9.55DD305 pKa = 3.21TLTSGDD311 pKa = 3.72RR312 pKa = 11.84FWRR315 pKa = 11.84YY316 pKa = 9.71SRR318 pKa = 11.84GIDD321 pKa = 3.15PGFFLPLQQSKK332 pKa = 10.89CYY334 pKa = 10.39ILIARR339 pKa = 11.84MADD342 pKa = 2.76ILVRR346 pKa = 11.84SGIVSVTQYY355 pKa = 11.13SDD357 pKa = 3.18PRR359 pKa = 11.84NARR362 pKa = 11.84CIADD366 pKa = 3.54KK367 pKa = 8.94TTVKK371 pKa = 10.56AQAEE375 pKa = 4.37IFGKK379 pKa = 9.04EE380 pKa = 3.4FWAAYY385 pKa = 9.79SSLSGSSADD394 pKa = 3.56SGPVAKK400 pKa = 9.72TIASGSRR407 pKa = 11.84TGNITVKK414 pKa = 10.64RR415 pKa = 11.84NLHH418 pKa = 6.0PRR420 pKa = 11.84APPPAPVVNMPPPQSSAPGALDD442 pKa = 3.76EE443 pKa = 5.13MVDD446 pKa = 4.42DD447 pKa = 6.78DD448 pKa = 4.56EE449 pKa = 4.95AQQ451 pKa = 3.07
Molecular weight: 48.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6AWM7|A0A1W6AWM7_9RHAB Movement protein OS=Citrus leprosis virus N OX=1956177 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.29SASSSSTLSEE11 pKa = 4.13EE12 pKa = 4.27CTVMSVRR19 pKa = 11.84SRR21 pKa = 11.84ITITSTEE28 pKa = 4.29KK29 pKa = 10.31IVPGPDD35 pKa = 2.48ITAKK39 pKa = 10.3IIQSLSLVNSKK50 pKa = 10.74AKK52 pKa = 10.77APDD55 pKa = 3.96GEE57 pKa = 4.63DD58 pKa = 3.46VQGDD62 pKa = 3.67WAQMLEE68 pKa = 4.37EE69 pKa = 4.19VAKK72 pKa = 10.64LSKK75 pKa = 10.05PQISGPFVDD84 pKa = 5.42KK85 pKa = 10.97DD86 pKa = 3.61LEE88 pKa = 4.38RR89 pKa = 11.84GNIYY93 pKa = 9.92SLNMIIGGILDD104 pKa = 3.95SQSGRR109 pKa = 11.84AGKK112 pKa = 10.75VMTLRR117 pKa = 11.84TGGLYY122 pKa = 7.68DD123 pKa = 3.55TPNYY127 pKa = 9.57HH128 pKa = 6.59INGRR132 pKa = 11.84SLAPKK137 pKa = 9.49INKK140 pKa = 8.88DD141 pKa = 3.09ASLEE145 pKa = 4.0ITATVATVKK154 pKa = 10.6RR155 pKa = 11.84PVPLCKK161 pKa = 10.01QKK163 pKa = 10.44IQDD166 pKa = 4.03YY167 pKa = 10.42IILLKK172 pKa = 10.83VVINPPTTGRR182 pKa = 11.84GRR184 pKa = 11.84SGG186 pKa = 2.94
MM1 pKa = 7.29SASSSSTLSEE11 pKa = 4.13EE12 pKa = 4.27CTVMSVRR19 pKa = 11.84SRR21 pKa = 11.84ITITSTEE28 pKa = 4.29KK29 pKa = 10.31IVPGPDD35 pKa = 2.48ITAKK39 pKa = 10.3IIQSLSLVNSKK50 pKa = 10.74AKK52 pKa = 10.77APDD55 pKa = 3.96GEE57 pKa = 4.63DD58 pKa = 3.46VQGDD62 pKa = 3.67WAQMLEE68 pKa = 4.37EE69 pKa = 4.19VAKK72 pKa = 10.64LSKK75 pKa = 10.05PQISGPFVDD84 pKa = 5.42KK85 pKa = 10.97DD86 pKa = 3.61LEE88 pKa = 4.38RR89 pKa = 11.84GNIYY93 pKa = 9.92SLNMIIGGILDD104 pKa = 3.95SQSGRR109 pKa = 11.84AGKK112 pKa = 10.75VMTLRR117 pKa = 11.84TGGLYY122 pKa = 7.68DD123 pKa = 3.55TPNYY127 pKa = 9.57HH128 pKa = 6.59INGRR132 pKa = 11.84SLAPKK137 pKa = 9.49INKK140 pKa = 8.88DD141 pKa = 3.09ASLEE145 pKa = 4.0ITATVATVKK154 pKa = 10.6RR155 pKa = 11.84PVPLCKK161 pKa = 10.01QKK163 pKa = 10.44IQDD166 pKa = 4.03YY167 pKa = 10.42IILLKK172 pKa = 10.83VVINPPTTGRR182 pKa = 11.84GRR184 pKa = 11.84SGG186 pKa = 2.94
Molecular weight: 19.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3606 |
186 |
1879 |
601.0 |
67.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.491 ± 0.994 | 1.886 ± 0.149 |
5.824 ± 0.214 | 5.241 ± 0.396 |
2.884 ± 0.32 | 6.378 ± 0.604 |
2.94 ± 0.34 | 7.404 ± 0.386 |
5.574 ± 0.511 | 8.735 ± 0.507 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.966 ± 0.149 | 3.966 ± 0.308 |
4.853 ± 0.575 | 2.856 ± 0.431 |
5.186 ± 0.719 | 8.93 ± 0.449 |
6.378 ± 0.56 | 6.517 ± 0.358 |
1.581 ± 0.206 | 3.411 ± 0.42 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
