
Hubei sobemo-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.86
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
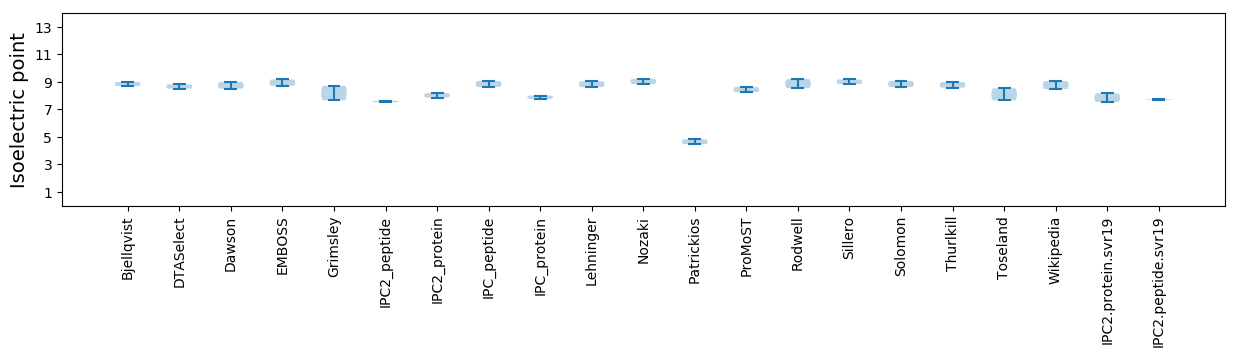
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE41|A0A1L3KE41_9VIRU C2H2-type domain-containing protein OS=Hubei sobemo-like virus 18 OX=1923203 PE=4 SV=1
MM1 pKa = 7.45RR2 pKa = 11.84TMIMQNFLDD11 pKa = 3.74LHH13 pKa = 6.81RR14 pKa = 11.84DD15 pKa = 3.51ARR17 pKa = 11.84WDD19 pKa = 3.5LDD21 pKa = 3.52EE22 pKa = 6.58DD23 pKa = 4.16FDD25 pKa = 4.82SFEE28 pKa = 4.2GFMRR32 pKa = 11.84SLKK35 pKa = 10.68RR36 pKa = 11.84IDD38 pKa = 4.15LSSNPGYY45 pKa = 10.08PYY47 pKa = 10.16MRR49 pKa = 11.84GCTSNRR55 pKa = 11.84QMLCYY60 pKa = 10.17KK61 pKa = 10.58NSDD64 pKa = 3.23QFDD67 pKa = 4.16PNKK70 pKa = 10.17VSILWDD76 pKa = 3.11IVRR79 pKa = 11.84QKK81 pKa = 11.39LNGSGADD88 pKa = 3.86PIKK91 pKa = 10.86LFVKK95 pKa = 10.05PEE97 pKa = 3.75PHH99 pKa = 6.54KK100 pKa = 10.61KK101 pKa = 10.38SKK103 pKa = 10.49IEE105 pKa = 3.79EE106 pKa = 3.71KK107 pKa = 10.11RR108 pKa = 11.84YY109 pKa = 10.25RR110 pKa = 11.84LISSVSLADD119 pKa = 3.39QLVDD123 pKa = 3.39HH124 pKa = 6.43MLFGKK129 pKa = 10.06MNDD132 pKa = 3.48KK133 pKa = 10.99LIASWPYY140 pKa = 10.53LPSKK144 pKa = 10.29VGWSFIGGGWKK155 pKa = 8.92TMPKK159 pKa = 9.78GGSWLAIDD167 pKa = 4.19KK168 pKa = 10.86SSWDD172 pKa = 3.16WTVQLWLLDD181 pKa = 3.71LVLQARR187 pKa = 11.84YY188 pKa = 8.59EE189 pKa = 4.16LCSTRR194 pKa = 11.84GEE196 pKa = 4.54VVDD199 pKa = 4.66RR200 pKa = 11.84WLKK203 pKa = 7.61MASMRR208 pKa = 11.84YY209 pKa = 8.39RR210 pKa = 11.84QLFKK214 pKa = 10.95EE215 pKa = 4.7PIFITSGGLLLKK227 pKa = 10.03QKK229 pKa = 10.3RR230 pKa = 11.84HH231 pKa = 4.98GVQKK235 pKa = 10.26SGCVNTIADD244 pKa = 3.81NSMMQSLLHH253 pKa = 6.35IRR255 pKa = 11.84VCLEE259 pKa = 3.81TKK261 pKa = 10.31QSVGVLYY268 pKa = 11.35SMGDD272 pKa = 3.42DD273 pKa = 3.82TLQQAPEE280 pKa = 4.04RR281 pKa = 11.84QNDD284 pKa = 3.74YY285 pKa = 11.4LSVLSKK291 pKa = 11.08YY292 pKa = 10.69CIIKK296 pKa = 10.24QADD299 pKa = 3.72ALVEE303 pKa = 3.96FAGFRR308 pKa = 11.84FDD310 pKa = 3.59GMRR313 pKa = 11.84VEE315 pKa = 4.25PVYY318 pKa = 10.62HH319 pKa = 6.59GKK321 pKa = 9.14HH322 pKa = 5.67AFNLLHH328 pKa = 7.45ADD330 pKa = 3.74PKK332 pKa = 10.95VLSDD336 pKa = 3.87LAKK339 pKa = 10.34SYY341 pKa = 10.61PLLYY345 pKa = 10.19HH346 pKa = 6.92RR347 pKa = 11.84SSNRR351 pKa = 11.84AWFRR355 pKa = 11.84EE356 pKa = 4.2LFLDD360 pKa = 3.54WHH362 pKa = 6.63GEE364 pKa = 4.23VPSIDD369 pKa = 4.05YY370 pKa = 10.78LDD372 pKa = 4.51SIYY375 pKa = 11.09DD376 pKa = 3.52GFEE379 pKa = 3.57
MM1 pKa = 7.45RR2 pKa = 11.84TMIMQNFLDD11 pKa = 3.74LHH13 pKa = 6.81RR14 pKa = 11.84DD15 pKa = 3.51ARR17 pKa = 11.84WDD19 pKa = 3.5LDD21 pKa = 3.52EE22 pKa = 6.58DD23 pKa = 4.16FDD25 pKa = 4.82SFEE28 pKa = 4.2GFMRR32 pKa = 11.84SLKK35 pKa = 10.68RR36 pKa = 11.84IDD38 pKa = 4.15LSSNPGYY45 pKa = 10.08PYY47 pKa = 10.16MRR49 pKa = 11.84GCTSNRR55 pKa = 11.84QMLCYY60 pKa = 10.17KK61 pKa = 10.58NSDD64 pKa = 3.23QFDD67 pKa = 4.16PNKK70 pKa = 10.17VSILWDD76 pKa = 3.11IVRR79 pKa = 11.84QKK81 pKa = 11.39LNGSGADD88 pKa = 3.86PIKK91 pKa = 10.86LFVKK95 pKa = 10.05PEE97 pKa = 3.75PHH99 pKa = 6.54KK100 pKa = 10.61KK101 pKa = 10.38SKK103 pKa = 10.49IEE105 pKa = 3.79EE106 pKa = 3.71KK107 pKa = 10.11RR108 pKa = 11.84YY109 pKa = 10.25RR110 pKa = 11.84LISSVSLADD119 pKa = 3.39QLVDD123 pKa = 3.39HH124 pKa = 6.43MLFGKK129 pKa = 10.06MNDD132 pKa = 3.48KK133 pKa = 10.99LIASWPYY140 pKa = 10.53LPSKK144 pKa = 10.29VGWSFIGGGWKK155 pKa = 8.92TMPKK159 pKa = 9.78GGSWLAIDD167 pKa = 4.19KK168 pKa = 10.86SSWDD172 pKa = 3.16WTVQLWLLDD181 pKa = 3.71LVLQARR187 pKa = 11.84YY188 pKa = 8.59EE189 pKa = 4.16LCSTRR194 pKa = 11.84GEE196 pKa = 4.54VVDD199 pKa = 4.66RR200 pKa = 11.84WLKK203 pKa = 7.61MASMRR208 pKa = 11.84YY209 pKa = 8.39RR210 pKa = 11.84QLFKK214 pKa = 10.95EE215 pKa = 4.7PIFITSGGLLLKK227 pKa = 10.03QKK229 pKa = 10.3RR230 pKa = 11.84HH231 pKa = 4.98GVQKK235 pKa = 10.26SGCVNTIADD244 pKa = 3.81NSMMQSLLHH253 pKa = 6.35IRR255 pKa = 11.84VCLEE259 pKa = 3.81TKK261 pKa = 10.31QSVGVLYY268 pKa = 11.35SMGDD272 pKa = 3.42DD273 pKa = 3.82TLQQAPEE280 pKa = 4.04RR281 pKa = 11.84QNDD284 pKa = 3.74YY285 pKa = 11.4LSVLSKK291 pKa = 11.08YY292 pKa = 10.69CIIKK296 pKa = 10.24QADD299 pKa = 3.72ALVEE303 pKa = 3.96FAGFRR308 pKa = 11.84FDD310 pKa = 3.59GMRR313 pKa = 11.84VEE315 pKa = 4.25PVYY318 pKa = 10.62HH319 pKa = 6.59GKK321 pKa = 9.14HH322 pKa = 5.67AFNLLHH328 pKa = 7.45ADD330 pKa = 3.74PKK332 pKa = 10.95VLSDD336 pKa = 3.87LAKK339 pKa = 10.34SYY341 pKa = 10.61PLLYY345 pKa = 10.19HH346 pKa = 6.92RR347 pKa = 11.84SSNRR351 pKa = 11.84AWFRR355 pKa = 11.84EE356 pKa = 4.2LFLDD360 pKa = 3.54WHH362 pKa = 6.63GEE364 pKa = 4.23VPSIDD369 pKa = 4.05YY370 pKa = 10.78LDD372 pKa = 4.51SIYY375 pKa = 11.09DD376 pKa = 3.52GFEE379 pKa = 3.57
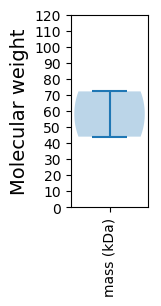
Molecular weight: 43.95 kDa
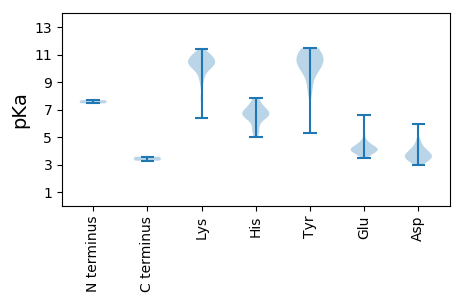
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE41|A0A1L3KE41_9VIRU C2H2-type domain-containing protein OS=Hubei sobemo-like virus 18 OX=1923203 PE=4 SV=1
MM1 pKa = 7.68EE2 pKa = 5.24PQPQVFPVEE11 pKa = 4.02EE12 pKa = 4.25GVYY15 pKa = 10.57CEE17 pKa = 4.07PTLDD21 pKa = 4.17GFVEE25 pKa = 4.71KK26 pKa = 10.1IWAKK30 pKa = 9.27IKK32 pKa = 10.56KK33 pKa = 8.79VSLVIWRR40 pKa = 11.84VWLLLVHH47 pKa = 6.72AWLYY51 pKa = 8.47YY52 pKa = 5.27TTKK55 pKa = 10.8DD56 pKa = 3.33YY57 pKa = 11.23FKK59 pKa = 10.68EE60 pKa = 4.14KK61 pKa = 10.38EE62 pKa = 4.2EE63 pKa = 4.18KK64 pKa = 10.27PEE66 pKa = 3.58VWFPRR71 pKa = 11.84LRR73 pKa = 11.84NQFNDD78 pKa = 3.09LSSILFKK85 pKa = 11.15LSLTFDD91 pKa = 4.21LEE93 pKa = 4.24WFVLVVTTTSLAFLTFVFVIKK114 pKa = 10.56RR115 pKa = 11.84SRR117 pKa = 11.84KK118 pKa = 6.4TLMMIRR124 pKa = 11.84GVRR127 pKa = 11.84FEE129 pKa = 4.53AMVEE133 pKa = 3.97GSDD136 pKa = 4.16FVKK139 pKa = 10.93GDD141 pKa = 3.15IPKK144 pKa = 10.02YY145 pKa = 9.56QVAVMKK151 pKa = 10.69AGLLRR156 pKa = 11.84DD157 pKa = 3.44VHH159 pKa = 7.82VGFGIRR165 pKa = 11.84VNNFLVVPTHH175 pKa = 6.81VINEE179 pKa = 4.13VGNAMILKK187 pKa = 9.73KK188 pKa = 10.31KK189 pKa = 10.41IKK191 pKa = 9.47IQVSMGQRR199 pKa = 11.84IISDD203 pKa = 3.77YY204 pKa = 11.4VSDD207 pKa = 4.07LTYY210 pKa = 11.08LPISNQIWADD220 pKa = 3.57LGVTKK225 pKa = 10.89SNLAKK230 pKa = 10.57EE231 pKa = 4.56SLNLAAMATIVGNGGQTTGLLRR253 pKa = 11.84KK254 pKa = 9.41CAIPFFYY261 pKa = 9.55TYY263 pKa = 10.58SASTVPGMSGAAYY276 pKa = 9.76AINNIVHH283 pKa = 7.58AIHH286 pKa = 6.63CGVVGEE292 pKa = 4.32NNSAISTVVVNKK304 pKa = 9.92EE305 pKa = 3.44LSKK308 pKa = 10.67IMKK311 pKa = 9.76GEE313 pKa = 3.9AVGATSISSNVDD325 pKa = 3.24MSWLPDD331 pKa = 3.63VEE333 pKa = 5.3NVWNDD338 pKa = 4.39DD339 pKa = 3.78LFEE342 pKa = 5.67DD343 pKa = 4.8YY344 pKa = 10.99NDD346 pKa = 3.71KK347 pKa = 10.95KK348 pKa = 10.74EE349 pKa = 4.01KK350 pKa = 10.4RR351 pKa = 11.84LIAARR356 pKa = 11.84EE357 pKa = 3.93KK358 pKa = 10.99HH359 pKa = 5.42GDD361 pKa = 3.3KK362 pKa = 10.97YY363 pKa = 11.45NPPTSGWSAMMDD375 pKa = 3.39YY376 pKa = 11.12DD377 pKa = 5.72DD378 pKa = 5.96NFAEE382 pKa = 4.55QYY384 pKa = 10.63GEE386 pKa = 4.07SSSGSKK392 pKa = 10.45SSLNVGKK399 pKa = 10.13NKK401 pKa = 10.5SPVKK405 pKa = 10.81VRR407 pKa = 11.84ITPQNNTDD415 pKa = 3.59TEE417 pKa = 4.54SEE419 pKa = 4.1IEE421 pKa = 4.65VYY423 pKa = 9.97VHH425 pKa = 6.84PADD428 pKa = 4.64AKK430 pKa = 9.65PTTKK434 pKa = 10.42SQLEE438 pKa = 4.2DD439 pKa = 3.87KK440 pKa = 10.66ISRR443 pKa = 11.84TEE445 pKa = 3.9NGSLEE450 pKa = 4.03NLHH453 pKa = 6.89LRR455 pKa = 11.84IQRR458 pKa = 11.84LEE460 pKa = 4.06GEE462 pKa = 4.29VFKK465 pKa = 10.77IAKK468 pKa = 9.43CQVCGMWVNQLSKK481 pKa = 10.45HH482 pKa = 5.54VKK484 pKa = 10.11DD485 pKa = 3.3MHH487 pKa = 6.35KK488 pKa = 10.89EE489 pKa = 3.87KK490 pKa = 10.28MSYY493 pKa = 10.34PCGKK497 pKa = 9.84CSAICRR503 pKa = 11.84SKK505 pKa = 11.15EE506 pKa = 3.63KK507 pKa = 10.36LASHH511 pKa = 6.56MLSHH515 pKa = 7.27PEE517 pKa = 3.63RR518 pKa = 11.84PKK520 pKa = 10.71CDD522 pKa = 2.94RR523 pKa = 11.84CGGFFRR529 pKa = 11.84RR530 pKa = 11.84EE531 pKa = 3.72EE532 pKa = 3.88NLVNHH537 pKa = 7.14RR538 pKa = 11.84MTCLPKK544 pKa = 10.31VRR546 pKa = 11.84FNARR550 pKa = 11.84NEE552 pKa = 4.0EE553 pKa = 4.27VVVVGEE559 pKa = 4.16SAIPSDD565 pKa = 3.87FKK567 pKa = 11.11TSVKK571 pKa = 9.99VDD573 pKa = 3.0KK574 pKa = 10.95KK575 pKa = 11.18DD576 pKa = 3.6FLGKK580 pKa = 10.18QGSQRR585 pKa = 11.84KK586 pKa = 7.89NLRR589 pKa = 11.84SSKK592 pKa = 8.97KK593 pKa = 7.86TSIASGEE600 pKa = 4.05KK601 pKa = 10.38DD602 pKa = 3.4LLTSQLEE609 pKa = 4.35FQSAMLKK616 pKa = 10.19SQDD619 pKa = 3.9AMLEE623 pKa = 4.03LLRR626 pKa = 11.84GLALGTNGQNLVTSQNN642 pKa = 3.28
MM1 pKa = 7.68EE2 pKa = 5.24PQPQVFPVEE11 pKa = 4.02EE12 pKa = 4.25GVYY15 pKa = 10.57CEE17 pKa = 4.07PTLDD21 pKa = 4.17GFVEE25 pKa = 4.71KK26 pKa = 10.1IWAKK30 pKa = 9.27IKK32 pKa = 10.56KK33 pKa = 8.79VSLVIWRR40 pKa = 11.84VWLLLVHH47 pKa = 6.72AWLYY51 pKa = 8.47YY52 pKa = 5.27TTKK55 pKa = 10.8DD56 pKa = 3.33YY57 pKa = 11.23FKK59 pKa = 10.68EE60 pKa = 4.14KK61 pKa = 10.38EE62 pKa = 4.2EE63 pKa = 4.18KK64 pKa = 10.27PEE66 pKa = 3.58VWFPRR71 pKa = 11.84LRR73 pKa = 11.84NQFNDD78 pKa = 3.09LSSILFKK85 pKa = 11.15LSLTFDD91 pKa = 4.21LEE93 pKa = 4.24WFVLVVTTTSLAFLTFVFVIKK114 pKa = 10.56RR115 pKa = 11.84SRR117 pKa = 11.84KK118 pKa = 6.4TLMMIRR124 pKa = 11.84GVRR127 pKa = 11.84FEE129 pKa = 4.53AMVEE133 pKa = 3.97GSDD136 pKa = 4.16FVKK139 pKa = 10.93GDD141 pKa = 3.15IPKK144 pKa = 10.02YY145 pKa = 9.56QVAVMKK151 pKa = 10.69AGLLRR156 pKa = 11.84DD157 pKa = 3.44VHH159 pKa = 7.82VGFGIRR165 pKa = 11.84VNNFLVVPTHH175 pKa = 6.81VINEE179 pKa = 4.13VGNAMILKK187 pKa = 9.73KK188 pKa = 10.31KK189 pKa = 10.41IKK191 pKa = 9.47IQVSMGQRR199 pKa = 11.84IISDD203 pKa = 3.77YY204 pKa = 11.4VSDD207 pKa = 4.07LTYY210 pKa = 11.08LPISNQIWADD220 pKa = 3.57LGVTKK225 pKa = 10.89SNLAKK230 pKa = 10.57EE231 pKa = 4.56SLNLAAMATIVGNGGQTTGLLRR253 pKa = 11.84KK254 pKa = 9.41CAIPFFYY261 pKa = 9.55TYY263 pKa = 10.58SASTVPGMSGAAYY276 pKa = 9.76AINNIVHH283 pKa = 7.58AIHH286 pKa = 6.63CGVVGEE292 pKa = 4.32NNSAISTVVVNKK304 pKa = 9.92EE305 pKa = 3.44LSKK308 pKa = 10.67IMKK311 pKa = 9.76GEE313 pKa = 3.9AVGATSISSNVDD325 pKa = 3.24MSWLPDD331 pKa = 3.63VEE333 pKa = 5.3NVWNDD338 pKa = 4.39DD339 pKa = 3.78LFEE342 pKa = 5.67DD343 pKa = 4.8YY344 pKa = 10.99NDD346 pKa = 3.71KK347 pKa = 10.95KK348 pKa = 10.74EE349 pKa = 4.01KK350 pKa = 10.4RR351 pKa = 11.84LIAARR356 pKa = 11.84EE357 pKa = 3.93KK358 pKa = 10.99HH359 pKa = 5.42GDD361 pKa = 3.3KK362 pKa = 10.97YY363 pKa = 11.45NPPTSGWSAMMDD375 pKa = 3.39YY376 pKa = 11.12DD377 pKa = 5.72DD378 pKa = 5.96NFAEE382 pKa = 4.55QYY384 pKa = 10.63GEE386 pKa = 4.07SSSGSKK392 pKa = 10.45SSLNVGKK399 pKa = 10.13NKK401 pKa = 10.5SPVKK405 pKa = 10.81VRR407 pKa = 11.84ITPQNNTDD415 pKa = 3.59TEE417 pKa = 4.54SEE419 pKa = 4.1IEE421 pKa = 4.65VYY423 pKa = 9.97VHH425 pKa = 6.84PADD428 pKa = 4.64AKK430 pKa = 9.65PTTKK434 pKa = 10.42SQLEE438 pKa = 4.2DD439 pKa = 3.87KK440 pKa = 10.66ISRR443 pKa = 11.84TEE445 pKa = 3.9NGSLEE450 pKa = 4.03NLHH453 pKa = 6.89LRR455 pKa = 11.84IQRR458 pKa = 11.84LEE460 pKa = 4.06GEE462 pKa = 4.29VFKK465 pKa = 10.77IAKK468 pKa = 9.43CQVCGMWVNQLSKK481 pKa = 10.45HH482 pKa = 5.54VKK484 pKa = 10.11DD485 pKa = 3.3MHH487 pKa = 6.35KK488 pKa = 10.89EE489 pKa = 3.87KK490 pKa = 10.28MSYY493 pKa = 10.34PCGKK497 pKa = 9.84CSAICRR503 pKa = 11.84SKK505 pKa = 11.15EE506 pKa = 3.63KK507 pKa = 10.36LASHH511 pKa = 6.56MLSHH515 pKa = 7.27PEE517 pKa = 3.63RR518 pKa = 11.84PKK520 pKa = 10.71CDD522 pKa = 2.94RR523 pKa = 11.84CGGFFRR529 pKa = 11.84RR530 pKa = 11.84EE531 pKa = 3.72EE532 pKa = 3.88NLVNHH537 pKa = 7.14RR538 pKa = 11.84MTCLPKK544 pKa = 10.31VRR546 pKa = 11.84FNARR550 pKa = 11.84NEE552 pKa = 4.0EE553 pKa = 4.27VVVVGEE559 pKa = 4.16SAIPSDD565 pKa = 3.87FKK567 pKa = 11.11TSVKK571 pKa = 9.99VDD573 pKa = 3.0KK574 pKa = 10.95KK575 pKa = 11.18DD576 pKa = 3.6FLGKK580 pKa = 10.18QGSQRR585 pKa = 11.84KK586 pKa = 7.89NLRR589 pKa = 11.84SSKK592 pKa = 8.97KK593 pKa = 7.86TSIASGEE600 pKa = 4.05KK601 pKa = 10.38DD602 pKa = 3.4LLTSQLEE609 pKa = 4.35FQSAMLKK616 pKa = 10.19SQDD619 pKa = 3.9AMLEE623 pKa = 4.03LLRR626 pKa = 11.84GLALGTNGQNLVTSQNN642 pKa = 3.28
Molecular weight: 72.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1021 |
379 |
642 |
510.5 |
58.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.093 ± 0.491 | 1.665 ± 0.046 |
5.877 ± 1.148 | 5.583 ± 0.915 |
4.212 ± 0.154 | 6.17 ± 0.091 |
2.253 ± 0.217 | 4.995 ± 0.138 |
8.227 ± 0.621 | 9.696 ± 1.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.428 ± 0.298 | 4.701 ± 0.864 |
3.722 ± 0.133 | 3.624 ± 0.337 |
4.995 ± 0.605 | 8.619 ± 0.198 |
4.016 ± 0.924 | 7.835 ± 1.292 |
2.253 ± 0.514 | 3.036 ± 0.519 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
