
Winogradskyella sp. KYW1333
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella; unclassified Winogradskyella
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
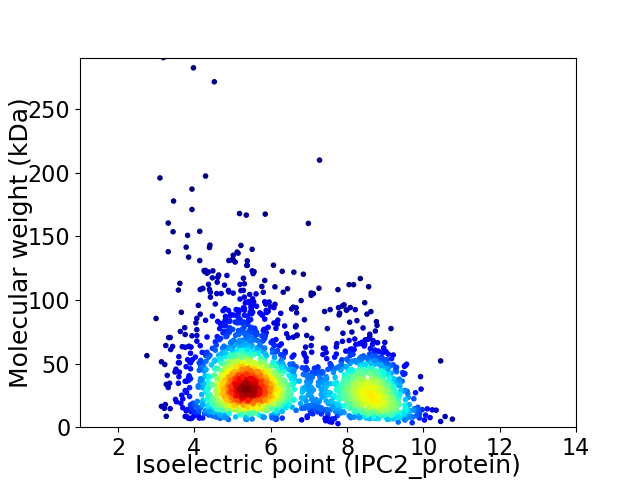
Virtual 2D-PAGE plot for 2480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A368MDQ9|A0A368MDQ9_9FLAO Low molecular weight phosphotyrosine protein phosphatase OS=Winogradskyella sp. KYW1333 OX=2282123 GN=DUZ96_09670 PE=3 SV=1
MM1 pKa = 7.66KK2 pKa = 10.16FYY4 pKa = 10.97QLTLYY9 pKa = 10.44LLSINLSVAQIAFDD23 pKa = 4.19NEE25 pKa = 4.09AEE27 pKa = 4.28TLGVEE32 pKa = 4.66IICGNTLFGNGVSFFDD48 pKa = 4.22YY49 pKa = 11.27DD50 pKa = 3.47NDD52 pKa = 3.52GWDD55 pKa = 4.19DD56 pKa = 3.95LTVATADD63 pKa = 3.79DD64 pKa = 4.36DD65 pKa = 4.01PVRR68 pKa = 11.84FYY70 pKa = 11.87KK71 pKa = 10.66NINGNFVEE79 pKa = 4.33QTLNIANNNWRR90 pKa = 11.84NKK92 pKa = 7.23QVNWVDD98 pKa = 3.06IDD100 pKa = 3.84NDD102 pKa = 3.57GDD104 pKa = 3.59NDD106 pKa = 4.35LFVTSDD112 pKa = 3.06TSINRR117 pKa = 11.84LYY119 pKa = 11.02EE120 pKa = 3.84NLGNMILQDD129 pKa = 3.35ITSTSGLLNEE139 pKa = 4.3VLVTYY144 pKa = 9.26GASWGDD150 pKa = 3.61YY151 pKa = 10.85NNDD154 pKa = 2.74GFLDD158 pKa = 3.77VFLSNRR164 pKa = 11.84DD165 pKa = 3.4TTIPNILYY173 pKa = 10.41KK174 pKa = 11.14NNGNNTFSLVNSEE187 pKa = 4.61SGLITTGTLSFCSAFIDD204 pKa = 3.95YY205 pKa = 10.98NKK207 pKa = 10.95DD208 pKa = 3.05GFQDD212 pKa = 3.3IYY214 pKa = 10.95VANDD218 pKa = 2.87KK219 pKa = 11.1VEE221 pKa = 4.22YY222 pKa = 9.54PNIMYY227 pKa = 10.48KK228 pKa = 10.85NNGDD232 pKa = 3.65GTFTEE237 pKa = 4.82VGFEE241 pKa = 4.03TGTDD245 pKa = 3.13ITIDD249 pKa = 3.47AMSVTIGDD257 pKa = 4.05YY258 pKa = 11.34NSDD261 pKa = 3.52SWLDD265 pKa = 3.28IYY267 pKa = 9.62VTNDD271 pKa = 2.85VVGNVLLKK279 pKa = 11.16NNGNGTFSDD288 pKa = 3.8EE289 pKa = 4.35TLSSNTAMNSVAWGSVFLDD308 pKa = 3.71AEE310 pKa = 4.23NDD312 pKa = 3.57GDD314 pKa = 4.22LDD316 pKa = 3.79IYY318 pKa = 11.37VSAEE322 pKa = 3.55HH323 pKa = 7.24DD324 pKa = 3.94GTVSGYY330 pKa = 10.69LSSALFTNNNDD341 pKa = 3.09EE342 pKa = 4.65YY343 pKa = 10.71ILEE346 pKa = 4.39NEE348 pKa = 5.39AIPDD352 pKa = 4.08DD353 pKa = 4.1YY354 pKa = 11.55AISYY358 pKa = 10.51SNAIGDD364 pKa = 4.2IDD366 pKa = 4.12NDD368 pKa = 4.46GYY370 pKa = 10.44PEE372 pKa = 3.9ILVNNINHH380 pKa = 6.97DD381 pKa = 5.11NIFLWKK387 pKa = 10.51NNSNNTNNWLKK398 pKa = 10.99VKK400 pKa = 11.03LEE402 pKa = 3.99GTISNRR408 pKa = 11.84NGVGSMIEE416 pKa = 3.55ISVNGEE422 pKa = 3.36KK423 pKa = 10.22QYY425 pKa = 11.04RR426 pKa = 11.84YY427 pKa = 7.6TLIGEE432 pKa = 5.23GYY434 pKa = 10.25LSQNSASEE442 pKa = 3.84IFGIGDD448 pKa = 3.53ATVIDD453 pKa = 4.18YY454 pKa = 11.05VKK456 pKa = 10.94VNWLSGTEE464 pKa = 4.0DD465 pKa = 2.44IFYY468 pKa = 10.87NVSPNQNLKK477 pKa = 9.89IVEE480 pKa = 4.24GSSLSIEE487 pKa = 4.2EE488 pKa = 4.76SNTSEE493 pKa = 3.95IKK495 pKa = 10.65LFPIPTTEE503 pKa = 3.55ILNIEE508 pKa = 4.01SSLPIEE514 pKa = 4.06EE515 pKa = 4.07VMVYY519 pKa = 10.93NLLGEE524 pKa = 4.44VNISANMNKK533 pKa = 9.71NEE535 pKa = 3.85ISLDD539 pKa = 3.74VTSLEE544 pKa = 3.78PGIYY548 pKa = 10.11FLRR551 pKa = 11.84LSLNSSQEE559 pKa = 3.56IIKK562 pKa = 9.88FIKK565 pKa = 9.96KK566 pKa = 9.87
MM1 pKa = 7.66KK2 pKa = 10.16FYY4 pKa = 10.97QLTLYY9 pKa = 10.44LLSINLSVAQIAFDD23 pKa = 4.19NEE25 pKa = 4.09AEE27 pKa = 4.28TLGVEE32 pKa = 4.66IICGNTLFGNGVSFFDD48 pKa = 4.22YY49 pKa = 11.27DD50 pKa = 3.47NDD52 pKa = 3.52GWDD55 pKa = 4.19DD56 pKa = 3.95LTVATADD63 pKa = 3.79DD64 pKa = 4.36DD65 pKa = 4.01PVRR68 pKa = 11.84FYY70 pKa = 11.87KK71 pKa = 10.66NINGNFVEE79 pKa = 4.33QTLNIANNNWRR90 pKa = 11.84NKK92 pKa = 7.23QVNWVDD98 pKa = 3.06IDD100 pKa = 3.84NDD102 pKa = 3.57GDD104 pKa = 3.59NDD106 pKa = 4.35LFVTSDD112 pKa = 3.06TSINRR117 pKa = 11.84LYY119 pKa = 11.02EE120 pKa = 3.84NLGNMILQDD129 pKa = 3.35ITSTSGLLNEE139 pKa = 4.3VLVTYY144 pKa = 9.26GASWGDD150 pKa = 3.61YY151 pKa = 10.85NNDD154 pKa = 2.74GFLDD158 pKa = 3.77VFLSNRR164 pKa = 11.84DD165 pKa = 3.4TTIPNILYY173 pKa = 10.41KK174 pKa = 11.14NNGNNTFSLVNSEE187 pKa = 4.61SGLITTGTLSFCSAFIDD204 pKa = 3.95YY205 pKa = 10.98NKK207 pKa = 10.95DD208 pKa = 3.05GFQDD212 pKa = 3.3IYY214 pKa = 10.95VANDD218 pKa = 2.87KK219 pKa = 11.1VEE221 pKa = 4.22YY222 pKa = 9.54PNIMYY227 pKa = 10.48KK228 pKa = 10.85NNGDD232 pKa = 3.65GTFTEE237 pKa = 4.82VGFEE241 pKa = 4.03TGTDD245 pKa = 3.13ITIDD249 pKa = 3.47AMSVTIGDD257 pKa = 4.05YY258 pKa = 11.34NSDD261 pKa = 3.52SWLDD265 pKa = 3.28IYY267 pKa = 9.62VTNDD271 pKa = 2.85VVGNVLLKK279 pKa = 11.16NNGNGTFSDD288 pKa = 3.8EE289 pKa = 4.35TLSSNTAMNSVAWGSVFLDD308 pKa = 3.71AEE310 pKa = 4.23NDD312 pKa = 3.57GDD314 pKa = 4.22LDD316 pKa = 3.79IYY318 pKa = 11.37VSAEE322 pKa = 3.55HH323 pKa = 7.24DD324 pKa = 3.94GTVSGYY330 pKa = 10.69LSSALFTNNNDD341 pKa = 3.09EE342 pKa = 4.65YY343 pKa = 10.71ILEE346 pKa = 4.39NEE348 pKa = 5.39AIPDD352 pKa = 4.08DD353 pKa = 4.1YY354 pKa = 11.55AISYY358 pKa = 10.51SNAIGDD364 pKa = 4.2IDD366 pKa = 4.12NDD368 pKa = 4.46GYY370 pKa = 10.44PEE372 pKa = 3.9ILVNNINHH380 pKa = 6.97DD381 pKa = 5.11NIFLWKK387 pKa = 10.51NNSNNTNNWLKK398 pKa = 10.99VKK400 pKa = 11.03LEE402 pKa = 3.99GTISNRR408 pKa = 11.84NGVGSMIEE416 pKa = 3.55ISVNGEE422 pKa = 3.36KK423 pKa = 10.22QYY425 pKa = 11.04RR426 pKa = 11.84YY427 pKa = 7.6TLIGEE432 pKa = 5.23GYY434 pKa = 10.25LSQNSASEE442 pKa = 3.84IFGIGDD448 pKa = 3.53ATVIDD453 pKa = 4.18YY454 pKa = 11.05VKK456 pKa = 10.94VNWLSGTEE464 pKa = 4.0DD465 pKa = 2.44IFYY468 pKa = 10.87NVSPNQNLKK477 pKa = 9.89IVEE480 pKa = 4.24GSSLSIEE487 pKa = 4.2EE488 pKa = 4.76SNTSEE493 pKa = 3.95IKK495 pKa = 10.65LFPIPTTEE503 pKa = 3.55ILNIEE508 pKa = 4.01SSLPIEE514 pKa = 4.06EE515 pKa = 4.07VMVYY519 pKa = 10.93NLLGEE524 pKa = 4.44VNISANMNKK533 pKa = 9.71NEE535 pKa = 3.85ISLDD539 pKa = 3.74VTSLEE544 pKa = 3.78PGIYY548 pKa = 10.11FLRR551 pKa = 11.84LSLNSSQEE559 pKa = 3.56IIKK562 pKa = 9.88FIKK565 pKa = 9.96KK566 pKa = 9.87
Molecular weight: 62.86 kDa
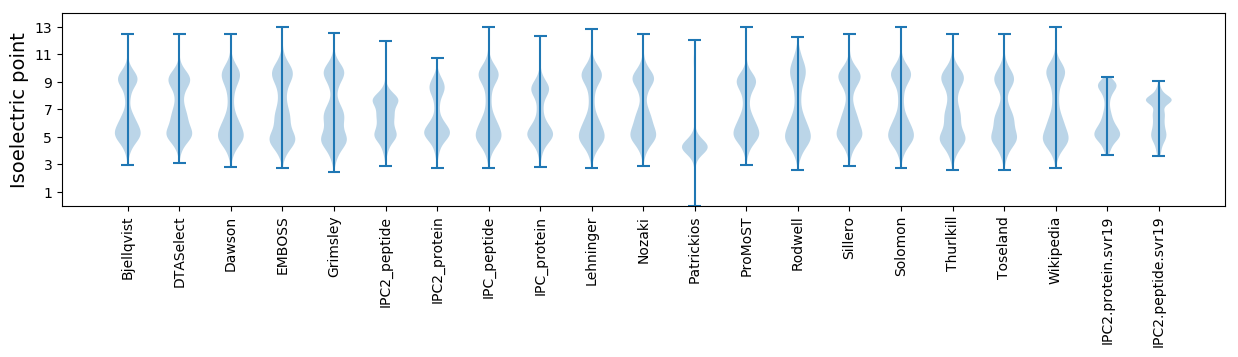
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A368MIZ7|A0A368MIZ7_9FLAO Glycoside hydrolase family 16 protein OS=Winogradskyella sp. KYW1333 OX=2282123 GN=DUZ96_01740 PE=3 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
859683 |
23 |
2737 |
346.6 |
39.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.99 ± 0.051 | 0.755 ± 0.014 |
5.989 ± 0.04 | 6.54 ± 0.044 |
5.277 ± 0.044 | 6.456 ± 0.046 |
1.712 ± 0.023 | 8.244 ± 0.039 |
7.479 ± 0.066 | 9.159 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.142 ± 0.025 | 6.552 ± 0.052 |
3.318 ± 0.031 | 3.185 ± 0.026 |
3.494 ± 0.033 | 6.663 ± 0.041 |
5.677 ± 0.047 | 6.203 ± 0.036 |
1.045 ± 0.019 | 4.119 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
