
Avon-Heathcote Estuary associated circular virus 25
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
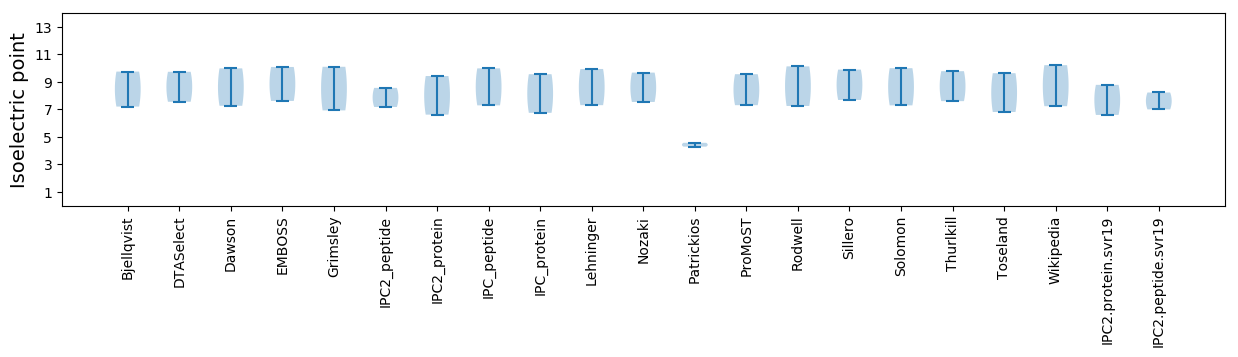
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
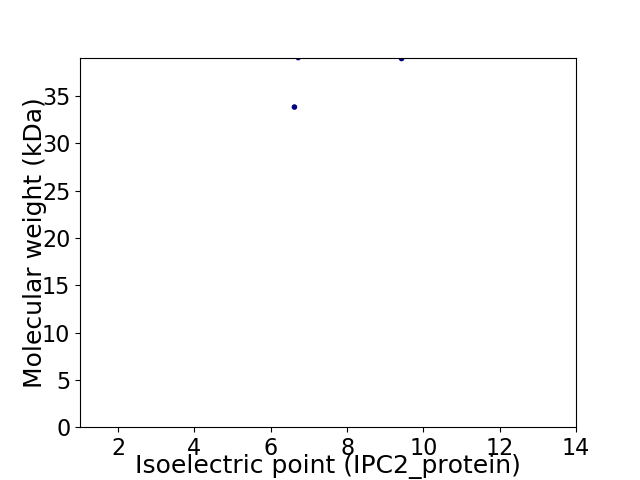
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBT8|A0A0C5IBT8_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 25 OX=1618249 PE=4 SV=1
MM1 pKa = 7.25ARR3 pKa = 11.84EE4 pKa = 3.8NRR6 pKa = 11.84FRR8 pKa = 11.84LQCRR12 pKa = 11.84RR13 pKa = 11.84LFLTYY18 pKa = 7.92PHH20 pKa = 6.75CTASIQEE27 pKa = 4.42VYY29 pKa = 11.2DD30 pKa = 4.68LIHH33 pKa = 6.51SKK35 pKa = 11.04KK36 pKa = 9.28EE37 pKa = 3.5IKK39 pKa = 9.62FARR42 pKa = 11.84ICIEE46 pKa = 3.85PHH48 pKa = 6.42SDD50 pKa = 3.29GTPHH54 pKa = 5.88IHH56 pKa = 6.34AAVMFAKK63 pKa = 10.14KK64 pKa = 10.77VNICNARR71 pKa = 11.84LFDD74 pKa = 5.69INDD77 pKa = 3.31HH78 pKa = 6.08HH79 pKa = 8.36CNMEE83 pKa = 4.77SIKK86 pKa = 10.61NWPATLNYY94 pKa = 9.39CKK96 pKa = 10.53KK97 pKa = 10.2GDD99 pKa = 3.42NWEE102 pKa = 5.1DD103 pKa = 3.43FDD105 pKa = 6.37NDD107 pKa = 4.47DD108 pKa = 6.22DD109 pKa = 5.43PDD111 pKa = 4.33QPMQDD116 pKa = 3.31NLFEE120 pKa = 4.46LAEE123 pKa = 4.28TMPSNDD129 pKa = 2.35FWEE132 pKa = 4.35YY133 pKa = 11.06CRR135 pKa = 11.84LKK137 pKa = 10.52KK138 pKa = 10.95VPFAYY143 pKa = 10.2ASNAMRR149 pKa = 11.84KK150 pKa = 8.2KK151 pKa = 10.64KK152 pKa = 10.93SMFTITDD159 pKa = 3.44STPYY163 pKa = 10.75GIIRR167 pKa = 11.84EE168 pKa = 4.13DD169 pKa = 3.13LGAYY173 pKa = 7.83TITEE177 pKa = 4.4DD178 pKa = 4.86SPATLASTVLVGPSGIGKK196 pKa = 7.24TSWAIRR202 pKa = 11.84EE203 pKa = 4.23SEE205 pKa = 4.29KK206 pKa = 10.73PSLLVTHH213 pKa = 6.75MDD215 pKa = 3.95ALRR218 pKa = 11.84KK219 pKa = 10.13LNGSHH224 pKa = 7.06KK225 pKa = 10.75SIIFDD230 pKa = 3.66DD231 pKa = 3.8MSFTHH236 pKa = 7.1IPRR239 pKa = 11.84EE240 pKa = 4.27AQIHH244 pKa = 5.26LVDD247 pKa = 4.17RR248 pKa = 11.84FQPRR252 pKa = 11.84QVHH255 pKa = 4.64VRR257 pKa = 11.84YY258 pKa = 8.23GTADD262 pKa = 3.13IPAGIQKK269 pKa = 9.79IFTANQYY276 pKa = 10.75PFVADD281 pKa = 3.47PAIDD285 pKa = 3.03RR286 pKa = 11.84RR287 pKa = 11.84VHH289 pKa = 6.12NKK291 pKa = 9.56NLYY294 pKa = 9.31
MM1 pKa = 7.25ARR3 pKa = 11.84EE4 pKa = 3.8NRR6 pKa = 11.84FRR8 pKa = 11.84LQCRR12 pKa = 11.84RR13 pKa = 11.84LFLTYY18 pKa = 7.92PHH20 pKa = 6.75CTASIQEE27 pKa = 4.42VYY29 pKa = 11.2DD30 pKa = 4.68LIHH33 pKa = 6.51SKK35 pKa = 11.04KK36 pKa = 9.28EE37 pKa = 3.5IKK39 pKa = 9.62FARR42 pKa = 11.84ICIEE46 pKa = 3.85PHH48 pKa = 6.42SDD50 pKa = 3.29GTPHH54 pKa = 5.88IHH56 pKa = 6.34AAVMFAKK63 pKa = 10.14KK64 pKa = 10.77VNICNARR71 pKa = 11.84LFDD74 pKa = 5.69INDD77 pKa = 3.31HH78 pKa = 6.08HH79 pKa = 8.36CNMEE83 pKa = 4.77SIKK86 pKa = 10.61NWPATLNYY94 pKa = 9.39CKK96 pKa = 10.53KK97 pKa = 10.2GDD99 pKa = 3.42NWEE102 pKa = 5.1DD103 pKa = 3.43FDD105 pKa = 6.37NDD107 pKa = 4.47DD108 pKa = 6.22DD109 pKa = 5.43PDD111 pKa = 4.33QPMQDD116 pKa = 3.31NLFEE120 pKa = 4.46LAEE123 pKa = 4.28TMPSNDD129 pKa = 2.35FWEE132 pKa = 4.35YY133 pKa = 11.06CRR135 pKa = 11.84LKK137 pKa = 10.52KK138 pKa = 10.95VPFAYY143 pKa = 10.2ASNAMRR149 pKa = 11.84KK150 pKa = 8.2KK151 pKa = 10.64KK152 pKa = 10.93SMFTITDD159 pKa = 3.44STPYY163 pKa = 10.75GIIRR167 pKa = 11.84EE168 pKa = 4.13DD169 pKa = 3.13LGAYY173 pKa = 7.83TITEE177 pKa = 4.4DD178 pKa = 4.86SPATLASTVLVGPSGIGKK196 pKa = 7.24TSWAIRR202 pKa = 11.84EE203 pKa = 4.23SEE205 pKa = 4.29KK206 pKa = 10.73PSLLVTHH213 pKa = 6.75MDD215 pKa = 3.95ALRR218 pKa = 11.84KK219 pKa = 10.13LNGSHH224 pKa = 7.06KK225 pKa = 10.75SIIFDD230 pKa = 3.66DD231 pKa = 3.8MSFTHH236 pKa = 7.1IPRR239 pKa = 11.84EE240 pKa = 4.27AQIHH244 pKa = 5.26LVDD247 pKa = 4.17RR248 pKa = 11.84FQPRR252 pKa = 11.84QVHH255 pKa = 4.64VRR257 pKa = 11.84YY258 pKa = 8.23GTADD262 pKa = 3.13IPAGIQKK269 pKa = 9.79IFTANQYY276 pKa = 10.75PFVADD281 pKa = 3.47PAIDD285 pKa = 3.03RR286 pKa = 11.84RR287 pKa = 11.84VHH289 pKa = 6.12NKK291 pKa = 9.56NLYY294 pKa = 9.31
Molecular weight: 33.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBT8|A0A0C5IBT8_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 25 OX=1618249 PE=4 SV=1
MM1 pKa = 7.69SYY3 pKa = 10.58NGRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VARR11 pKa = 11.84SSRR14 pKa = 11.84YY15 pKa = 7.48NARR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 10.19SPYY23 pKa = 10.15SPYY26 pKa = 10.91SLRR29 pKa = 11.84SAVRR33 pKa = 11.84VGRR36 pKa = 11.84YY37 pKa = 8.57LGNYY41 pKa = 8.01ARR43 pKa = 11.84KK44 pKa = 10.01SFNNWRR50 pKa = 11.84SKK52 pKa = 9.87TSAKK56 pKa = 9.47KK57 pKa = 8.88QRR59 pKa = 11.84RR60 pKa = 11.84NIGNKK65 pKa = 9.47IISASNGIGYY75 pKa = 10.1SYY77 pKa = 10.68FRR79 pKa = 11.84EE80 pKa = 4.0NRR82 pKa = 11.84KK83 pKa = 9.88CYY85 pKa = 9.69PNVRR89 pKa = 11.84NMLKK93 pKa = 10.51AGTLSVYY100 pKa = 10.38KK101 pKa = 10.88YY102 pKa = 10.94VDD104 pKa = 3.42TKK106 pKa = 10.89RR107 pKa = 11.84LEE109 pKa = 3.94WDD111 pKa = 3.19AGSQGVLNVATTFGVQQIQNVMNNIGASANGDD143 pKa = 3.28DD144 pKa = 4.09TTRR147 pKa = 11.84MVVKK151 pKa = 10.48GVTGSVNISNMSNANVLITLYY172 pKa = 10.89DD173 pKa = 3.5VVARR177 pKa = 11.84TDD179 pKa = 3.8SNTSSAWANPLNAWGNGLEE198 pKa = 4.22DD199 pKa = 3.9SEE201 pKa = 5.04EE202 pKa = 4.51GGSVAAGRR210 pKa = 11.84NNIGATPFQSVMFCRR225 pKa = 11.84KK226 pKa = 8.25YY227 pKa = 10.89RR228 pKa = 11.84VMKK231 pKa = 8.15VTKK234 pKa = 9.56IYY236 pKa = 9.7MEE238 pKa = 4.55AGRR241 pKa = 11.84SHH243 pKa = 5.78IHH245 pKa = 5.39YY246 pKa = 9.96IKK248 pKa = 10.83RR249 pKa = 11.84NMNSVLNNSIMEE261 pKa = 4.32TNEE264 pKa = 3.48TNLANKK270 pKa = 7.55THH272 pKa = 6.23YY273 pKa = 10.16CFAVARR279 pKa = 11.84GFPVNDD285 pKa = 3.19ATTDD289 pKa = 3.42TNIALGSGALDD300 pKa = 3.39FVFTEE305 pKa = 5.61TYY307 pKa = 8.55QQVWPQYY314 pKa = 9.03ALKK317 pKa = 10.1RR318 pKa = 11.84YY319 pKa = 9.18YY320 pKa = 11.31YY321 pKa = 10.61NDD323 pKa = 3.18QQQAITSEE331 pKa = 3.97KK332 pKa = 10.63LMNAEE337 pKa = 4.01TGEE340 pKa = 4.03GDD342 pKa = 3.57VYY344 pKa = 11.54EE345 pKa = 4.43EE346 pKa = 4.31VV347 pKa = 3.4
MM1 pKa = 7.69SYY3 pKa = 10.58NGRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VARR11 pKa = 11.84SSRR14 pKa = 11.84YY15 pKa = 7.48NARR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 10.19SPYY23 pKa = 10.15SPYY26 pKa = 10.91SLRR29 pKa = 11.84SAVRR33 pKa = 11.84VGRR36 pKa = 11.84YY37 pKa = 8.57LGNYY41 pKa = 8.01ARR43 pKa = 11.84KK44 pKa = 10.01SFNNWRR50 pKa = 11.84SKK52 pKa = 9.87TSAKK56 pKa = 9.47KK57 pKa = 8.88QRR59 pKa = 11.84RR60 pKa = 11.84NIGNKK65 pKa = 9.47IISASNGIGYY75 pKa = 10.1SYY77 pKa = 10.68FRR79 pKa = 11.84EE80 pKa = 4.0NRR82 pKa = 11.84KK83 pKa = 9.88CYY85 pKa = 9.69PNVRR89 pKa = 11.84NMLKK93 pKa = 10.51AGTLSVYY100 pKa = 10.38KK101 pKa = 10.88YY102 pKa = 10.94VDD104 pKa = 3.42TKK106 pKa = 10.89RR107 pKa = 11.84LEE109 pKa = 3.94WDD111 pKa = 3.19AGSQGVLNVATTFGVQQIQNVMNNIGASANGDD143 pKa = 3.28DD144 pKa = 4.09TTRR147 pKa = 11.84MVVKK151 pKa = 10.48GVTGSVNISNMSNANVLITLYY172 pKa = 10.89DD173 pKa = 3.5VVARR177 pKa = 11.84TDD179 pKa = 3.8SNTSSAWANPLNAWGNGLEE198 pKa = 4.22DD199 pKa = 3.9SEE201 pKa = 5.04EE202 pKa = 4.51GGSVAAGRR210 pKa = 11.84NNIGATPFQSVMFCRR225 pKa = 11.84KK226 pKa = 8.25YY227 pKa = 10.89RR228 pKa = 11.84VMKK231 pKa = 8.15VTKK234 pKa = 9.56IYY236 pKa = 9.7MEE238 pKa = 4.55AGRR241 pKa = 11.84SHH243 pKa = 5.78IHH245 pKa = 5.39YY246 pKa = 9.96IKK248 pKa = 10.83RR249 pKa = 11.84NMNSVLNNSIMEE261 pKa = 4.32TNEE264 pKa = 3.48TNLANKK270 pKa = 7.55THH272 pKa = 6.23YY273 pKa = 10.16CFAVARR279 pKa = 11.84GFPVNDD285 pKa = 3.19ATTDD289 pKa = 3.42TNIALGSGALDD300 pKa = 3.39FVFTEE305 pKa = 5.61TYY307 pKa = 8.55QQVWPQYY314 pKa = 9.03ALKK317 pKa = 10.1RR318 pKa = 11.84YY319 pKa = 9.18YY320 pKa = 11.31YY321 pKa = 10.61NDD323 pKa = 3.18QQQAITSEE331 pKa = 3.97KK332 pKa = 10.63LMNAEE337 pKa = 4.01TGEE340 pKa = 4.03GDD342 pKa = 3.57VYY344 pKa = 11.54EE345 pKa = 4.43EE346 pKa = 4.31VV347 pKa = 3.4
Molecular weight: 38.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
641 |
294 |
347 |
320.5 |
36.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.112 ± 0.157 | 1.56 ± 0.444 |
5.46 ± 1.279 | 4.368 ± 0.213 |
3.744 ± 0.735 | 5.616 ± 1.199 |
2.496 ± 1.043 | 5.928 ± 1.026 |
5.616 ± 0.458 | 5.304 ± 0.443 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.12 ± 0.032 | 8.268 ± 1.53 |
3.744 ± 1.104 | 3.276 ± 0.116 |
7.02 ± 0.486 | 7.332 ± 0.839 |
6.24 ± 0.248 | 6.24 ± 1.169 |
1.404 ± 0.024 | 5.148 ± 0.946 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
