
bacterium HR18
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
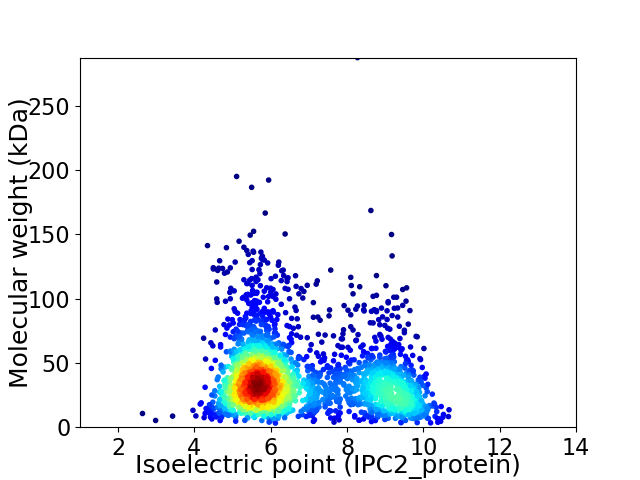
Virtual 2D-PAGE plot for 2437 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5XL89|A0A2H5XL89_9BACT D-inositol 3-phosphate glycosyltransferase OS=bacterium HR18 OX=2035413 GN=mshA_2 PE=4 SV=1
MM1 pKa = 7.49SFFDD5 pKa = 5.6FGFDD9 pKa = 4.08DD10 pKa = 5.77ADD12 pKa = 3.93DD13 pKa = 4.13ALEE16 pKa = 3.93RR17 pKa = 11.84HH18 pKa = 5.25QLEE21 pKa = 4.44EE22 pKa = 3.98LVAAYY27 pKa = 7.81EE28 pKa = 4.61AYY30 pKa = 10.31GSSAYY35 pKa = 10.06FDD37 pKa = 3.74SDD39 pKa = 3.43ALEE42 pKa = 4.9EE43 pKa = 4.15IATYY47 pKa = 10.51YY48 pKa = 10.14YY49 pKa = 9.8EE50 pKa = 3.73QGRR53 pKa = 11.84FEE55 pKa = 4.19EE56 pKa = 4.33ALGVIDD62 pKa = 5.89RR63 pKa = 11.84LLKK66 pKa = 10.08LHH68 pKa = 6.43PTASDD73 pKa = 2.24AWMRR77 pKa = 11.84RR78 pKa = 11.84GILLSHH84 pKa = 7.21LGQHH88 pKa = 5.95EE89 pKa = 4.48EE90 pKa = 4.05ALQAYY95 pKa = 7.78QQALTLNPVDD105 pKa = 3.94TEE107 pKa = 4.35TLVNLGITLDD117 pKa = 3.44NLGRR121 pKa = 11.84FEE123 pKa = 4.42EE124 pKa = 4.29ALEE127 pKa = 4.47AYY129 pKa = 8.21EE130 pKa = 5.19QALQLDD136 pKa = 4.37PLNEE140 pKa = 3.46EE141 pKa = 4.09AYY143 pKa = 9.01YY144 pKa = 11.02NRR146 pKa = 11.84GITLEE151 pKa = 3.9RR152 pKa = 11.84MDD154 pKa = 4.8RR155 pKa = 11.84LAEE158 pKa = 4.6AIAAFEE164 pKa = 3.87QAAALNPEE172 pKa = 4.42HH173 pKa = 7.1PEE175 pKa = 3.29VWYY178 pKa = 10.14EE179 pKa = 4.61LGYY182 pKa = 11.03CYY184 pKa = 10.69DD185 pKa = 3.8RR186 pKa = 11.84LGDD189 pKa = 3.98DD190 pKa = 4.02EE191 pKa = 6.28RR192 pKa = 11.84SLACYY197 pKa = 9.77DD198 pKa = 3.21RR199 pKa = 11.84HH200 pKa = 6.48LALDD204 pKa = 4.87PYY206 pKa = 11.26SADD209 pKa = 2.4AWYY212 pKa = 10.81NRR214 pKa = 11.84GIVLNRR220 pKa = 11.84MGRR223 pKa = 11.84YY224 pKa = 8.17EE225 pKa = 4.12EE226 pKa = 5.36AVASYY231 pKa = 10.93DD232 pKa = 3.55FAIAIQDD239 pKa = 3.91DD240 pKa = 4.09FASAYY245 pKa = 9.18YY246 pKa = 10.68NRR248 pKa = 11.84GNALTNLGDD257 pKa = 3.52LRR259 pKa = 11.84GAIEE263 pKa = 4.31SYY265 pKa = 10.85EE266 pKa = 3.99KK267 pKa = 10.84VLEE270 pKa = 4.18IEE272 pKa = 4.79GGDD275 pKa = 3.27PATYY279 pKa = 10.87YY280 pKa = 10.95NIALAYY286 pKa = 9.86EE287 pKa = 4.21EE288 pKa = 4.1LQEE291 pKa = 4.15YY292 pKa = 6.92EE293 pKa = 3.76TAIAYY298 pKa = 7.53FQQALEE304 pKa = 4.17EE305 pKa = 4.52DD306 pKa = 3.69PTYY309 pKa = 11.47AEE311 pKa = 3.47AWYY314 pKa = 10.45GLGCCYY320 pKa = 10.48DD321 pKa = 3.19ALEE324 pKa = 4.38RR325 pKa = 11.84FEE327 pKa = 4.92EE328 pKa = 4.53AVACLEE334 pKa = 4.07RR335 pKa = 11.84AVALRR340 pKa = 11.84PEE342 pKa = 4.52ASEE345 pKa = 3.84FWYY348 pKa = 10.51AKK350 pKa = 10.45ADD352 pKa = 3.52CEE354 pKa = 4.34YY355 pKa = 10.22NARR358 pKa = 11.84RR359 pKa = 11.84VEE361 pKa = 4.15AALHH365 pKa = 5.74SYY367 pKa = 9.9HH368 pKa = 6.78RR369 pKa = 11.84VVSLDD374 pKa = 3.24PTNRR378 pKa = 11.84EE379 pKa = 3.2AWLDD383 pKa = 3.57YY384 pKa = 11.4AEE386 pKa = 4.36TLFEE390 pKa = 4.41VGLTEE395 pKa = 4.22EE396 pKa = 4.15ALEE399 pKa = 4.69AYY401 pKa = 9.85RR402 pKa = 11.84QALSLNPDD410 pKa = 2.42ARR412 pKa = 11.84AYY414 pKa = 9.57IRR416 pKa = 11.84QARR419 pKa = 11.84ALLALGRR426 pKa = 11.84SEE428 pKa = 4.36EE429 pKa = 4.6GIRR432 pKa = 11.84SLKK435 pKa = 9.03MALRR439 pKa = 11.84LDD441 pKa = 4.14PSAKK445 pKa = 10.29DD446 pKa = 3.56EE447 pKa = 4.26LPEE450 pKa = 4.32FYY452 pKa = 10.1RR453 pKa = 11.84DD454 pKa = 3.08ASIRR458 pKa = 11.84RR459 pKa = 11.84QLGLDD464 pKa = 3.54GG465 pKa = 4.79
MM1 pKa = 7.49SFFDD5 pKa = 5.6FGFDD9 pKa = 4.08DD10 pKa = 5.77ADD12 pKa = 3.93DD13 pKa = 4.13ALEE16 pKa = 3.93RR17 pKa = 11.84HH18 pKa = 5.25QLEE21 pKa = 4.44EE22 pKa = 3.98LVAAYY27 pKa = 7.81EE28 pKa = 4.61AYY30 pKa = 10.31GSSAYY35 pKa = 10.06FDD37 pKa = 3.74SDD39 pKa = 3.43ALEE42 pKa = 4.9EE43 pKa = 4.15IATYY47 pKa = 10.51YY48 pKa = 10.14YY49 pKa = 9.8EE50 pKa = 3.73QGRR53 pKa = 11.84FEE55 pKa = 4.19EE56 pKa = 4.33ALGVIDD62 pKa = 5.89RR63 pKa = 11.84LLKK66 pKa = 10.08LHH68 pKa = 6.43PTASDD73 pKa = 2.24AWMRR77 pKa = 11.84RR78 pKa = 11.84GILLSHH84 pKa = 7.21LGQHH88 pKa = 5.95EE89 pKa = 4.48EE90 pKa = 4.05ALQAYY95 pKa = 7.78QQALTLNPVDD105 pKa = 3.94TEE107 pKa = 4.35TLVNLGITLDD117 pKa = 3.44NLGRR121 pKa = 11.84FEE123 pKa = 4.42EE124 pKa = 4.29ALEE127 pKa = 4.47AYY129 pKa = 8.21EE130 pKa = 5.19QALQLDD136 pKa = 4.37PLNEE140 pKa = 3.46EE141 pKa = 4.09AYY143 pKa = 9.01YY144 pKa = 11.02NRR146 pKa = 11.84GITLEE151 pKa = 3.9RR152 pKa = 11.84MDD154 pKa = 4.8RR155 pKa = 11.84LAEE158 pKa = 4.6AIAAFEE164 pKa = 3.87QAAALNPEE172 pKa = 4.42HH173 pKa = 7.1PEE175 pKa = 3.29VWYY178 pKa = 10.14EE179 pKa = 4.61LGYY182 pKa = 11.03CYY184 pKa = 10.69DD185 pKa = 3.8RR186 pKa = 11.84LGDD189 pKa = 3.98DD190 pKa = 4.02EE191 pKa = 6.28RR192 pKa = 11.84SLACYY197 pKa = 9.77DD198 pKa = 3.21RR199 pKa = 11.84HH200 pKa = 6.48LALDD204 pKa = 4.87PYY206 pKa = 11.26SADD209 pKa = 2.4AWYY212 pKa = 10.81NRR214 pKa = 11.84GIVLNRR220 pKa = 11.84MGRR223 pKa = 11.84YY224 pKa = 8.17EE225 pKa = 4.12EE226 pKa = 5.36AVASYY231 pKa = 10.93DD232 pKa = 3.55FAIAIQDD239 pKa = 3.91DD240 pKa = 4.09FASAYY245 pKa = 9.18YY246 pKa = 10.68NRR248 pKa = 11.84GNALTNLGDD257 pKa = 3.52LRR259 pKa = 11.84GAIEE263 pKa = 4.31SYY265 pKa = 10.85EE266 pKa = 3.99KK267 pKa = 10.84VLEE270 pKa = 4.18IEE272 pKa = 4.79GGDD275 pKa = 3.27PATYY279 pKa = 10.87YY280 pKa = 10.95NIALAYY286 pKa = 9.86EE287 pKa = 4.21EE288 pKa = 4.1LQEE291 pKa = 4.15YY292 pKa = 6.92EE293 pKa = 3.76TAIAYY298 pKa = 7.53FQQALEE304 pKa = 4.17EE305 pKa = 4.52DD306 pKa = 3.69PTYY309 pKa = 11.47AEE311 pKa = 3.47AWYY314 pKa = 10.45GLGCCYY320 pKa = 10.48DD321 pKa = 3.19ALEE324 pKa = 4.38RR325 pKa = 11.84FEE327 pKa = 4.92EE328 pKa = 4.53AVACLEE334 pKa = 4.07RR335 pKa = 11.84AVALRR340 pKa = 11.84PEE342 pKa = 4.52ASEE345 pKa = 3.84FWYY348 pKa = 10.51AKK350 pKa = 10.45ADD352 pKa = 3.52CEE354 pKa = 4.34YY355 pKa = 10.22NARR358 pKa = 11.84RR359 pKa = 11.84VEE361 pKa = 4.15AALHH365 pKa = 5.74SYY367 pKa = 9.9HH368 pKa = 6.78RR369 pKa = 11.84VVSLDD374 pKa = 3.24PTNRR378 pKa = 11.84EE379 pKa = 3.2AWLDD383 pKa = 3.57YY384 pKa = 11.4AEE386 pKa = 4.36TLFEE390 pKa = 4.41VGLTEE395 pKa = 4.22EE396 pKa = 4.15ALEE399 pKa = 4.69AYY401 pKa = 9.85RR402 pKa = 11.84QALSLNPDD410 pKa = 2.42ARR412 pKa = 11.84AYY414 pKa = 9.57IRR416 pKa = 11.84QARR419 pKa = 11.84ALLALGRR426 pKa = 11.84SEE428 pKa = 4.36EE429 pKa = 4.6GIRR432 pKa = 11.84SLKK435 pKa = 9.03MALRR439 pKa = 11.84LDD441 pKa = 4.14PSAKK445 pKa = 10.29DD446 pKa = 3.56EE447 pKa = 4.26LPEE450 pKa = 4.32FYY452 pKa = 10.1RR453 pKa = 11.84DD454 pKa = 3.08ASIRR458 pKa = 11.84RR459 pKa = 11.84QLGLDD464 pKa = 3.54GG465 pKa = 4.79
Molecular weight: 52.98 kDa
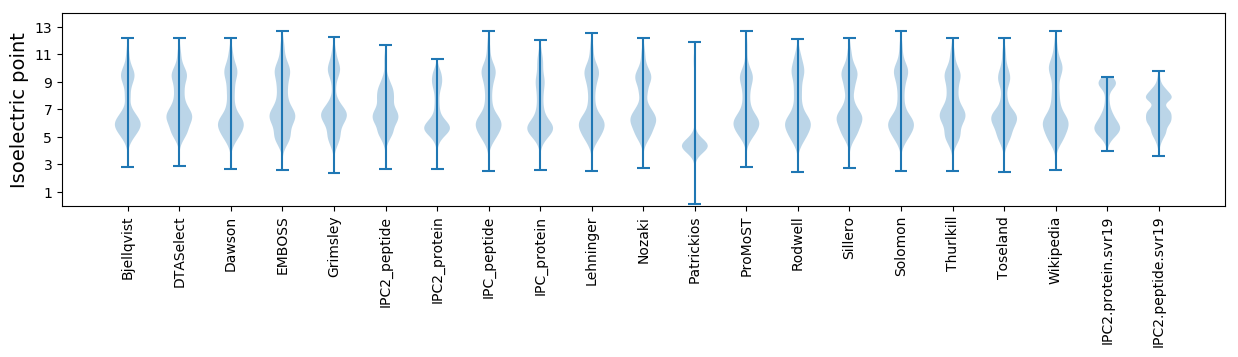
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5XMM0|A0A2H5XMM0_9BACT 30S ribosomal protein S19 OS=bacterium HR18 OX=2035413 GN=rpsS PE=3 SV=1
MM1 pKa = 7.37LVEE4 pKa = 4.05KK5 pKa = 10.51VRR7 pKa = 11.84RR8 pKa = 11.84FIEE11 pKa = 4.16LNGLLRR17 pKa = 11.84PGEE20 pKa = 4.04RR21 pKa = 11.84LLVGVSGGVDD31 pKa = 3.2SVVLLEE37 pKa = 4.09VLRR40 pKa = 11.84QLGYY44 pKa = 10.25SLIVAHH50 pKa = 6.34VNYY53 pKa = 9.96KK54 pKa = 10.59LRR56 pKa = 11.84GADD59 pKa = 3.27SDD61 pKa = 3.76ADD63 pKa = 3.84EE64 pKa = 4.86AFVRR68 pKa = 11.84ALCRR72 pKa = 11.84QYY74 pKa = 10.84RR75 pKa = 11.84IPIRR79 pKa = 11.84VARR82 pKa = 11.84LNLKK86 pKa = 10.25AKK88 pKa = 10.05VRR90 pKa = 11.84GGSIQAAARR99 pKa = 11.84SVRR102 pKa = 11.84YY103 pKa = 10.01AFFARR108 pKa = 11.84VAQRR112 pKa = 11.84EE113 pKa = 4.58GIEE116 pKa = 4.03AVAVGHH122 pKa = 6.28HH123 pKa = 6.23QDD125 pKa = 3.92DD126 pKa = 3.81QAEE129 pKa = 4.26TLLLNLLRR137 pKa = 11.84SSGLEE142 pKa = 3.89GLTGMAPVRR151 pKa = 11.84PFEE154 pKa = 4.3VGQKK158 pKa = 10.04LRR160 pKa = 11.84LVRR163 pKa = 11.84PLLSVTRR170 pKa = 11.84AEE172 pKa = 4.02IEE174 pKa = 3.62SWARR178 pKa = 11.84AKK180 pKa = 10.56GLSWRR185 pKa = 11.84EE186 pKa = 3.87DD187 pKa = 3.46VSNTSLNYY195 pKa = 9.58RR196 pKa = 11.84RR197 pKa = 11.84VLVRR201 pKa = 11.84QVVLPLLRR209 pKa = 11.84QHH211 pKa = 6.95FGPNVAEE218 pKa = 4.21RR219 pKa = 11.84LAHH222 pKa = 5.07TAEE225 pKa = 4.35LLRR228 pKa = 11.84AYY230 pKa = 7.74WQSTFAQVLMQHH242 pKa = 6.42WLSASEE248 pKa = 3.94SDD250 pKa = 4.25AVGGYY255 pKa = 9.53LRR257 pKa = 11.84QEE259 pKa = 4.49ALAGMPRR266 pKa = 11.84VWQQRR271 pKa = 11.84LILEE275 pKa = 4.23ALKK278 pKa = 10.35RR279 pKa = 11.84WLPGAPRR286 pKa = 11.84HH287 pKa = 6.5RR288 pKa = 11.84RR289 pKa = 11.84LAWQVLRR296 pKa = 11.84LLEE299 pKa = 4.29AQPGRR304 pKa = 11.84RR305 pKa = 11.84LKK307 pKa = 10.87LPQGTIWRR315 pKa = 11.84DD316 pKa = 3.08RR317 pKa = 11.84KK318 pKa = 9.67GLRR321 pKa = 11.84FRR323 pKa = 11.84RR324 pKa = 11.84CAPAAPISEE333 pKa = 4.34EE334 pKa = 4.1GALEE338 pKa = 4.1PGRR341 pKa = 11.84PLVLEE346 pKa = 4.34GGVFEE351 pKa = 6.05AVLLSQKK358 pKa = 9.21PACLNAGTPQVVYY371 pKa = 10.83VDD373 pKa = 4.05ADD375 pKa = 4.05RR376 pKa = 11.84LQWPLWVRR384 pKa = 11.84RR385 pKa = 11.84WRR387 pKa = 11.84PGDD390 pKa = 3.34RR391 pKa = 11.84LQPLGMQGHH400 pKa = 6.45KK401 pKa = 10.14KK402 pKa = 10.69VSDD405 pKa = 3.9LLTDD409 pKa = 3.64LRR411 pKa = 11.84VPVDD415 pKa = 3.99RR416 pKa = 11.84RR417 pKa = 11.84AHH419 pKa = 5.97SYY421 pKa = 8.57VVCQGEE427 pKa = 4.29EE428 pKa = 4.21IVWVAGYY435 pKa = 10.55RR436 pKa = 11.84LAEE439 pKa = 4.23PFRR442 pKa = 11.84VRR444 pKa = 11.84PEE446 pKa = 4.19TVHH449 pKa = 6.86IAQLQFTPIEE459 pKa = 4.28DD460 pKa = 3.33
MM1 pKa = 7.37LVEE4 pKa = 4.05KK5 pKa = 10.51VRR7 pKa = 11.84RR8 pKa = 11.84FIEE11 pKa = 4.16LNGLLRR17 pKa = 11.84PGEE20 pKa = 4.04RR21 pKa = 11.84LLVGVSGGVDD31 pKa = 3.2SVVLLEE37 pKa = 4.09VLRR40 pKa = 11.84QLGYY44 pKa = 10.25SLIVAHH50 pKa = 6.34VNYY53 pKa = 9.96KK54 pKa = 10.59LRR56 pKa = 11.84GADD59 pKa = 3.27SDD61 pKa = 3.76ADD63 pKa = 3.84EE64 pKa = 4.86AFVRR68 pKa = 11.84ALCRR72 pKa = 11.84QYY74 pKa = 10.84RR75 pKa = 11.84IPIRR79 pKa = 11.84VARR82 pKa = 11.84LNLKK86 pKa = 10.25AKK88 pKa = 10.05VRR90 pKa = 11.84GGSIQAAARR99 pKa = 11.84SVRR102 pKa = 11.84YY103 pKa = 10.01AFFARR108 pKa = 11.84VAQRR112 pKa = 11.84EE113 pKa = 4.58GIEE116 pKa = 4.03AVAVGHH122 pKa = 6.28HH123 pKa = 6.23QDD125 pKa = 3.92DD126 pKa = 3.81QAEE129 pKa = 4.26TLLLNLLRR137 pKa = 11.84SSGLEE142 pKa = 3.89GLTGMAPVRR151 pKa = 11.84PFEE154 pKa = 4.3VGQKK158 pKa = 10.04LRR160 pKa = 11.84LVRR163 pKa = 11.84PLLSVTRR170 pKa = 11.84AEE172 pKa = 4.02IEE174 pKa = 3.62SWARR178 pKa = 11.84AKK180 pKa = 10.56GLSWRR185 pKa = 11.84EE186 pKa = 3.87DD187 pKa = 3.46VSNTSLNYY195 pKa = 9.58RR196 pKa = 11.84RR197 pKa = 11.84VLVRR201 pKa = 11.84QVVLPLLRR209 pKa = 11.84QHH211 pKa = 6.95FGPNVAEE218 pKa = 4.21RR219 pKa = 11.84LAHH222 pKa = 5.07TAEE225 pKa = 4.35LLRR228 pKa = 11.84AYY230 pKa = 7.74WQSTFAQVLMQHH242 pKa = 6.42WLSASEE248 pKa = 3.94SDD250 pKa = 4.25AVGGYY255 pKa = 9.53LRR257 pKa = 11.84QEE259 pKa = 4.49ALAGMPRR266 pKa = 11.84VWQQRR271 pKa = 11.84LILEE275 pKa = 4.23ALKK278 pKa = 10.35RR279 pKa = 11.84WLPGAPRR286 pKa = 11.84HH287 pKa = 6.5RR288 pKa = 11.84RR289 pKa = 11.84LAWQVLRR296 pKa = 11.84LLEE299 pKa = 4.29AQPGRR304 pKa = 11.84RR305 pKa = 11.84LKK307 pKa = 10.87LPQGTIWRR315 pKa = 11.84DD316 pKa = 3.08RR317 pKa = 11.84KK318 pKa = 9.67GLRR321 pKa = 11.84FRR323 pKa = 11.84RR324 pKa = 11.84CAPAAPISEE333 pKa = 4.34EE334 pKa = 4.1GALEE338 pKa = 4.1PGRR341 pKa = 11.84PLVLEE346 pKa = 4.34GGVFEE351 pKa = 6.05AVLLSQKK358 pKa = 9.21PACLNAGTPQVVYY371 pKa = 10.83VDD373 pKa = 4.05ADD375 pKa = 4.05RR376 pKa = 11.84LQWPLWVRR384 pKa = 11.84RR385 pKa = 11.84WRR387 pKa = 11.84PGDD390 pKa = 3.34RR391 pKa = 11.84LQPLGMQGHH400 pKa = 6.45KK401 pKa = 10.14KK402 pKa = 10.69VSDD405 pKa = 3.9LLTDD409 pKa = 3.64LRR411 pKa = 11.84VPVDD415 pKa = 3.99RR416 pKa = 11.84RR417 pKa = 11.84AHH419 pKa = 5.97SYY421 pKa = 8.57VVCQGEE427 pKa = 4.29EE428 pKa = 4.21IVWVAGYY435 pKa = 10.55RR436 pKa = 11.84LAEE439 pKa = 4.23PFRR442 pKa = 11.84VRR444 pKa = 11.84PEE446 pKa = 4.19TVHH449 pKa = 6.86IAQLQFTPIEE459 pKa = 4.28DD460 pKa = 3.33
Molecular weight: 52.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
885271 |
29 |
2540 |
363.3 |
40.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.668 ± 0.061 | 0.756 ± 0.016 |
4.542 ± 0.034 | 6.5 ± 0.05 |
3.799 ± 0.031 | 7.507 ± 0.048 |
2.375 ± 0.024 | 4.481 ± 0.034 |
2.335 ± 0.032 | 12.087 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.854 ± 0.018 | 2.424 ± 0.034 |
5.751 ± 0.04 | 4.565 ± 0.038 |
8.144 ± 0.04 | 4.376 ± 0.032 |
5.321 ± 0.034 | 7.589 ± 0.038 |
1.658 ± 0.024 | 3.267 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
