
Wenling hoplichthys paramyxovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Hoplichthysvirus; Hoplichthys hoplichthysvirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
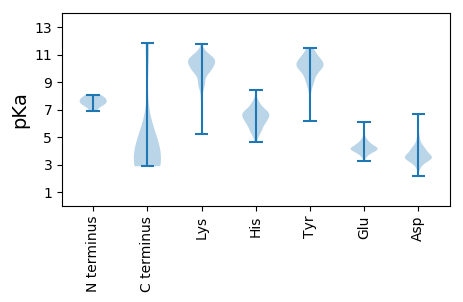
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2P1GNJ8|A0A2P1GNJ8_9MONO Fusion glycoprotein F0 OS=Wenling hoplichthys paramyxovirus OX=2116453 PE=3 SV=1
MM1 pKa = 7.43TMGTLVFMLLVLLAGEE17 pKa = 4.58CKK19 pKa = 10.66GGDD22 pKa = 3.91EE23 pKa = 4.51SCLYY27 pKa = 10.19PPALIQQEE35 pKa = 4.07GKK37 pKa = 9.88IFYY40 pKa = 10.21NGVPGTTRR48 pKa = 11.84LVSTDD53 pKa = 3.51RR54 pKa = 11.84VICPSTGCNIISDD67 pKa = 4.05GRR69 pKa = 11.84LSASADD75 pKa = 3.4AYY77 pKa = 10.96GSMDD81 pKa = 3.11MHH83 pKa = 6.83KK84 pKa = 10.24RR85 pKa = 11.84DD86 pKa = 3.36YY87 pKa = 11.19EE88 pKa = 4.22FYY90 pKa = 10.2KK91 pKa = 10.93DD92 pKa = 4.18KK93 pKa = 10.71IVSRR97 pKa = 11.84AGGALVANILRR108 pKa = 11.84TRR110 pKa = 11.84ANTALGFYY118 pKa = 9.23EE119 pKa = 5.71SCAEE123 pKa = 3.68WKK125 pKa = 10.08IFYY128 pKa = 9.85EE129 pKa = 4.57GVVFSPYY136 pKa = 9.12MRR138 pKa = 11.84QACTGTNGEE147 pKa = 4.09WSRR150 pKa = 11.84MKK152 pKa = 10.39EE153 pKa = 3.58QFYY156 pKa = 10.12TKK158 pKa = 9.62FYY160 pKa = 11.06EE161 pKa = 3.95MATRR165 pKa = 11.84FQCFKK170 pKa = 10.36FVRR173 pKa = 11.84VPYY176 pKa = 9.97IRR178 pKa = 11.84PAEE181 pKa = 4.22STQEE185 pKa = 3.95VAPGCGAGGTINGVEE200 pKa = 4.64YY201 pKa = 9.42ITDD204 pKa = 3.65PRR206 pKa = 11.84NGTIITSVVTDD217 pKa = 3.67LKK219 pKa = 10.46ITGEE223 pKa = 4.18YY224 pKa = 10.75LMPFCYY230 pKa = 9.33FVYY233 pKa = 10.23RR234 pKa = 11.84SRR236 pKa = 11.84DD237 pKa = 2.4IWLAEE242 pKa = 3.52NRR244 pKa = 11.84GYY246 pKa = 11.21DD247 pKa = 3.26SDD249 pKa = 4.66PFLKK253 pKa = 10.58VYY255 pKa = 10.72NEE257 pKa = 3.92QGEE260 pKa = 4.63TLFDD264 pKa = 3.75WNQLTDD270 pKa = 4.05GYY272 pKa = 11.1GEE274 pKa = 4.07PKK276 pKa = 9.61AEE278 pKa = 4.44KK279 pKa = 9.49MIKK282 pKa = 9.81CSFIIVEE289 pKa = 4.4KK290 pKa = 9.62TSKK293 pKa = 8.97ITIQSWSASQFITRR307 pKa = 11.84AAYY310 pKa = 9.78YY311 pKa = 10.2FNEE314 pKa = 4.76TIPRR318 pKa = 11.84RR319 pKa = 11.84PIQVIEE325 pKa = 4.48SKK327 pKa = 9.51WAQSRR332 pKa = 11.84MSDD335 pKa = 4.27PITQEE340 pKa = 3.86SACRR344 pKa = 11.84RR345 pKa = 11.84LGDD348 pKa = 3.28YY349 pKa = 10.72CIYY352 pKa = 10.55FRR354 pKa = 11.84IGTYY358 pKa = 10.44LGIFDD363 pKa = 5.86DD364 pKa = 4.74EE365 pKa = 5.43LGDD368 pKa = 3.75LQVRR372 pKa = 11.84DD373 pKa = 3.81ILSGQKK379 pKa = 10.41GGGPNFCGGRR389 pKa = 11.84SYY391 pKa = 11.23PGGCPASCNPSPFCTDD407 pKa = 4.03YY408 pKa = 11.37ISMGGATAHH417 pKa = 6.2MASPYY422 pKa = 10.05SSYY425 pKa = 11.2RR426 pKa = 11.84SFGSLYY432 pKa = 10.41EE433 pKa = 4.12KK434 pKa = 8.64WTLTYY439 pKa = 10.24PGGEE443 pKa = 4.21SVSTLLHH450 pKa = 5.46QLSLSMASEE459 pKa = 4.31WNTGSITPEE468 pKa = 4.09GEE470 pKa = 3.93PTLSLLGMDD479 pKa = 4.08SEE481 pKa = 4.37ISNFEE486 pKa = 3.93TLGYY490 pKa = 9.18TKK492 pKa = 10.65DD493 pKa = 3.51PEE495 pKa = 4.29TTRR498 pKa = 11.84WCVTLSCEE506 pKa = 3.97NCTNLFDD513 pKa = 4.04TRR515 pKa = 11.84TGRR518 pKa = 11.84PRR520 pKa = 11.84IAPFVASRR528 pKa = 11.84IKK530 pKa = 11.03AEE532 pKa = 3.95GLAEE536 pKa = 4.07IEE538 pKa = 5.0MRR540 pKa = 11.84DD541 pKa = 3.49YY542 pKa = 11.14WNDD545 pKa = 2.71VSLIIALCLLLALIIWLIVTSVLLTDD571 pKa = 3.29VRR573 pKa = 11.84RR574 pKa = 11.84TKK576 pKa = 10.29KK577 pKa = 10.03HH578 pKa = 5.51PRR580 pKa = 11.84SSIDD584 pKa = 3.18PSMEE588 pKa = 3.99GYY590 pKa = 10.3VADD593 pKa = 4.49LL594 pKa = 3.98
MM1 pKa = 7.43TMGTLVFMLLVLLAGEE17 pKa = 4.58CKK19 pKa = 10.66GGDD22 pKa = 3.91EE23 pKa = 4.51SCLYY27 pKa = 10.19PPALIQQEE35 pKa = 4.07GKK37 pKa = 9.88IFYY40 pKa = 10.21NGVPGTTRR48 pKa = 11.84LVSTDD53 pKa = 3.51RR54 pKa = 11.84VICPSTGCNIISDD67 pKa = 4.05GRR69 pKa = 11.84LSASADD75 pKa = 3.4AYY77 pKa = 10.96GSMDD81 pKa = 3.11MHH83 pKa = 6.83KK84 pKa = 10.24RR85 pKa = 11.84DD86 pKa = 3.36YY87 pKa = 11.19EE88 pKa = 4.22FYY90 pKa = 10.2KK91 pKa = 10.93DD92 pKa = 4.18KK93 pKa = 10.71IVSRR97 pKa = 11.84AGGALVANILRR108 pKa = 11.84TRR110 pKa = 11.84ANTALGFYY118 pKa = 9.23EE119 pKa = 5.71SCAEE123 pKa = 3.68WKK125 pKa = 10.08IFYY128 pKa = 9.85EE129 pKa = 4.57GVVFSPYY136 pKa = 9.12MRR138 pKa = 11.84QACTGTNGEE147 pKa = 4.09WSRR150 pKa = 11.84MKK152 pKa = 10.39EE153 pKa = 3.58QFYY156 pKa = 10.12TKK158 pKa = 9.62FYY160 pKa = 11.06EE161 pKa = 3.95MATRR165 pKa = 11.84FQCFKK170 pKa = 10.36FVRR173 pKa = 11.84VPYY176 pKa = 9.97IRR178 pKa = 11.84PAEE181 pKa = 4.22STQEE185 pKa = 3.95VAPGCGAGGTINGVEE200 pKa = 4.64YY201 pKa = 9.42ITDD204 pKa = 3.65PRR206 pKa = 11.84NGTIITSVVTDD217 pKa = 3.67LKK219 pKa = 10.46ITGEE223 pKa = 4.18YY224 pKa = 10.75LMPFCYY230 pKa = 9.33FVYY233 pKa = 10.23RR234 pKa = 11.84SRR236 pKa = 11.84DD237 pKa = 2.4IWLAEE242 pKa = 3.52NRR244 pKa = 11.84GYY246 pKa = 11.21DD247 pKa = 3.26SDD249 pKa = 4.66PFLKK253 pKa = 10.58VYY255 pKa = 10.72NEE257 pKa = 3.92QGEE260 pKa = 4.63TLFDD264 pKa = 3.75WNQLTDD270 pKa = 4.05GYY272 pKa = 11.1GEE274 pKa = 4.07PKK276 pKa = 9.61AEE278 pKa = 4.44KK279 pKa = 9.49MIKK282 pKa = 9.81CSFIIVEE289 pKa = 4.4KK290 pKa = 9.62TSKK293 pKa = 8.97ITIQSWSASQFITRR307 pKa = 11.84AAYY310 pKa = 9.78YY311 pKa = 10.2FNEE314 pKa = 4.76TIPRR318 pKa = 11.84RR319 pKa = 11.84PIQVIEE325 pKa = 4.48SKK327 pKa = 9.51WAQSRR332 pKa = 11.84MSDD335 pKa = 4.27PITQEE340 pKa = 3.86SACRR344 pKa = 11.84RR345 pKa = 11.84LGDD348 pKa = 3.28YY349 pKa = 10.72CIYY352 pKa = 10.55FRR354 pKa = 11.84IGTYY358 pKa = 10.44LGIFDD363 pKa = 5.86DD364 pKa = 4.74EE365 pKa = 5.43LGDD368 pKa = 3.75LQVRR372 pKa = 11.84DD373 pKa = 3.81ILSGQKK379 pKa = 10.41GGGPNFCGGRR389 pKa = 11.84SYY391 pKa = 11.23PGGCPASCNPSPFCTDD407 pKa = 4.03YY408 pKa = 11.37ISMGGATAHH417 pKa = 6.2MASPYY422 pKa = 10.05SSYY425 pKa = 11.2RR426 pKa = 11.84SFGSLYY432 pKa = 10.41EE433 pKa = 4.12KK434 pKa = 8.64WTLTYY439 pKa = 10.24PGGEE443 pKa = 4.21SVSTLLHH450 pKa = 5.46QLSLSMASEE459 pKa = 4.31WNTGSITPEE468 pKa = 4.09GEE470 pKa = 3.93PTLSLLGMDD479 pKa = 4.08SEE481 pKa = 4.37ISNFEE486 pKa = 3.93TLGYY490 pKa = 9.18TKK492 pKa = 10.65DD493 pKa = 3.51PEE495 pKa = 4.29TTRR498 pKa = 11.84WCVTLSCEE506 pKa = 3.97NCTNLFDD513 pKa = 4.04TRR515 pKa = 11.84TGRR518 pKa = 11.84PRR520 pKa = 11.84IAPFVASRR528 pKa = 11.84IKK530 pKa = 11.03AEE532 pKa = 3.95GLAEE536 pKa = 4.07IEE538 pKa = 5.0MRR540 pKa = 11.84DD541 pKa = 3.49YY542 pKa = 11.14WNDD545 pKa = 2.71VSLIIALCLLLALIIWLIVTSVLLTDD571 pKa = 3.29VRR573 pKa = 11.84RR574 pKa = 11.84TKK576 pKa = 10.29KK577 pKa = 10.03HH578 pKa = 5.51PRR580 pKa = 11.84SSIDD584 pKa = 3.18PSMEE588 pKa = 3.99GYY590 pKa = 10.3VADD593 pKa = 4.49LL594 pKa = 3.98
Molecular weight: 66.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GN21|A0A2P1GN21_9MONO Nucleocapsid OS=Wenling hoplichthys paramyxovirus OX=2116453 PE=3 SV=1
MM1 pKa = 7.72AARR4 pKa = 11.84EE5 pKa = 4.17LSRR8 pKa = 11.84AAYY11 pKa = 10.02DD12 pKa = 3.36HH13 pKa = 6.02EE14 pKa = 4.64TGIKK18 pKa = 10.41GPTDD22 pKa = 3.16GTKK25 pKa = 10.32CYY27 pKa = 7.48TTYY30 pKa = 11.06NPWGVGYY37 pKa = 7.67DD38 pKa = 3.4TSQISVFAIGDD49 pKa = 3.45IGKK52 pKa = 10.11DD53 pKa = 3.18KK54 pKa = 10.64ILKK57 pKa = 9.41NLRR60 pKa = 11.84INLGLLTEE68 pKa = 4.47GMTRR72 pKa = 11.84EE73 pKa = 3.9QAADD77 pKa = 4.95LITGTSFSAVKK88 pKa = 10.7VMGVNEE94 pKa = 4.79TIRR97 pKa = 11.84IKK99 pKa = 10.78PNSNMEE105 pKa = 4.39ALRR108 pKa = 11.84KK109 pKa = 8.51ITGLKK114 pKa = 9.72MGVQTALGPVCCPKK128 pKa = 10.76AYY130 pKa = 10.11VFPVGKK136 pKa = 9.9QLDD139 pKa = 3.61VRR141 pKa = 11.84FVHH144 pKa = 7.01VIFTVLPYY152 pKa = 10.56KK153 pKa = 10.8AGTHH157 pKa = 5.75KK158 pKa = 10.94LGGGILNLQKK168 pKa = 9.84TKK170 pKa = 10.4IVGGISVNVQTTGKK184 pKa = 8.52MGKK187 pKa = 8.82IDD189 pKa = 4.6LPLTIPFMEE198 pKa = 4.32WKK200 pKa = 10.21KK201 pKa = 8.74STEE204 pKa = 4.2GVFEE208 pKa = 4.39LMGMVMGASLKK219 pKa = 10.16VALGGVNGLSIHH231 pKa = 5.27VTAIGQIHH239 pKa = 7.44KK240 pKa = 9.81DD241 pKa = 3.48LKK243 pKa = 8.61GTGMNKK249 pKa = 9.84NMMLCPVFMYY259 pKa = 10.53PDD261 pKa = 3.31IVFSNCLKK269 pKa = 8.61MWKK272 pKa = 8.07VTSAKK277 pKa = 10.26LILQIQPRR285 pKa = 11.84VFRR288 pKa = 11.84PIEE291 pKa = 4.05EE292 pKa = 4.37PLVSEE297 pKa = 5.14RR298 pKa = 11.84DD299 pKa = 3.75LLTKK303 pKa = 10.26ALKK306 pKa = 10.74SGGEE310 pKa = 4.06EE311 pKa = 3.72EE312 pKa = 4.87DD313 pKa = 3.5
MM1 pKa = 7.72AARR4 pKa = 11.84EE5 pKa = 4.17LSRR8 pKa = 11.84AAYY11 pKa = 10.02DD12 pKa = 3.36HH13 pKa = 6.02EE14 pKa = 4.64TGIKK18 pKa = 10.41GPTDD22 pKa = 3.16GTKK25 pKa = 10.32CYY27 pKa = 7.48TTYY30 pKa = 11.06NPWGVGYY37 pKa = 7.67DD38 pKa = 3.4TSQISVFAIGDD49 pKa = 3.45IGKK52 pKa = 10.11DD53 pKa = 3.18KK54 pKa = 10.64ILKK57 pKa = 9.41NLRR60 pKa = 11.84INLGLLTEE68 pKa = 4.47GMTRR72 pKa = 11.84EE73 pKa = 3.9QAADD77 pKa = 4.95LITGTSFSAVKK88 pKa = 10.7VMGVNEE94 pKa = 4.79TIRR97 pKa = 11.84IKK99 pKa = 10.78PNSNMEE105 pKa = 4.39ALRR108 pKa = 11.84KK109 pKa = 8.51ITGLKK114 pKa = 9.72MGVQTALGPVCCPKK128 pKa = 10.76AYY130 pKa = 10.11VFPVGKK136 pKa = 9.9QLDD139 pKa = 3.61VRR141 pKa = 11.84FVHH144 pKa = 7.01VIFTVLPYY152 pKa = 10.56KK153 pKa = 10.8AGTHH157 pKa = 5.75KK158 pKa = 10.94LGGGILNLQKK168 pKa = 9.84TKK170 pKa = 10.4IVGGISVNVQTTGKK184 pKa = 8.52MGKK187 pKa = 8.82IDD189 pKa = 4.6LPLTIPFMEE198 pKa = 4.32WKK200 pKa = 10.21KK201 pKa = 8.74STEE204 pKa = 4.2GVFEE208 pKa = 4.39LMGMVMGASLKK219 pKa = 10.16VALGGVNGLSIHH231 pKa = 5.27VTAIGQIHH239 pKa = 7.44KK240 pKa = 9.81DD241 pKa = 3.48LKK243 pKa = 8.61GTGMNKK249 pKa = 9.84NMMLCPVFMYY259 pKa = 10.53PDD261 pKa = 3.31IVFSNCLKK269 pKa = 8.61MWKK272 pKa = 8.07VTSAKK277 pKa = 10.26LILQIQPRR285 pKa = 11.84VFRR288 pKa = 11.84PIEE291 pKa = 4.05EE292 pKa = 4.37PLVSEE297 pKa = 5.14RR298 pKa = 11.84DD299 pKa = 3.75LLTKK303 pKa = 10.26ALKK306 pKa = 10.74SGGEE310 pKa = 4.06EE311 pKa = 3.72EE312 pKa = 4.87DD313 pKa = 3.5
Molecular weight: 34.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4722 |
140 |
2055 |
524.7 |
58.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.612 ± 0.474 | 2.308 ± 0.333 |
5.04 ± 0.22 | 7.264 ± 0.622 |
3.346 ± 0.293 | 7.116 ± 0.46 |
1.588 ± 0.274 | 7.793 ± 0.444 |
6.777 ± 0.779 | 9.022 ± 0.464 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.283 ± 0.277 | 4.299 ± 0.401 |
3.685 ± 0.252 | 2.944 ± 0.239 |
5.548 ± 0.446 | 8.492 ± 0.563 |
6.501 ± 0.428 | 5.294 ± 0.437 |
1.207 ± 0.203 | 2.88 ± 0.42 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
