
Ophiocordyceps australis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Ophiocordycipitaceae; Ophiocordyceps
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
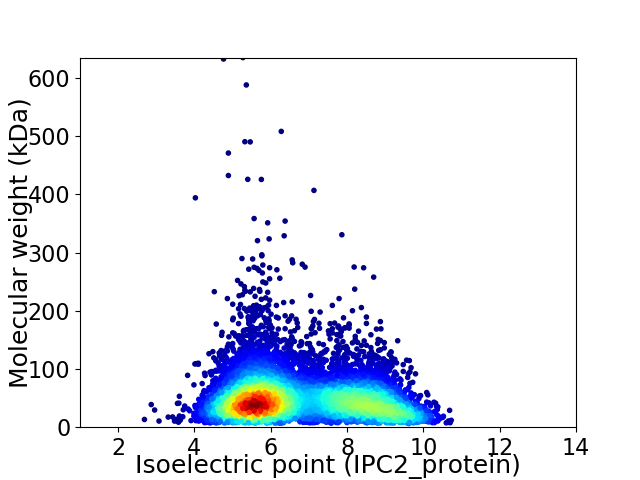
Virtual 2D-PAGE plot for 8168 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C5XHB8|A0A2C5XHB8_9HYPO Homeobox domain-containing protein OS=Ophiocordyceps australis OX=1399860 GN=CDD81_6872 PE=4 SV=1
MM1 pKa = 7.68SDD3 pKa = 3.05MAGEE7 pKa = 4.19LQPLGSQATSIAVDD21 pKa = 3.62EE22 pKa = 4.53FTKK25 pKa = 10.51FLKK28 pKa = 10.85NSVPTAMIPIDD39 pKa = 3.15IHH41 pKa = 5.91KK42 pKa = 10.36CLVYY46 pKa = 9.58TANCILDD53 pKa = 3.56VAKK56 pKa = 10.64EE57 pKa = 4.18SGVEE61 pKa = 3.8LSEE64 pKa = 3.69ITIGRR69 pKa = 11.84DD70 pKa = 4.05CMPTWPCCFWDD81 pKa = 3.78EE82 pKa = 4.5RR83 pKa = 11.84VEE85 pKa = 4.27TLHH88 pKa = 6.1EE89 pKa = 4.59TIQKK93 pKa = 10.13GLVVSQMVRR102 pKa = 11.84SLGDD106 pKa = 3.16FVRR109 pKa = 11.84LEE111 pKa = 4.48IINKK115 pKa = 9.73GNMKK119 pKa = 10.1LDD121 pKa = 3.7EE122 pKa = 4.43ARR124 pKa = 11.84KK125 pKa = 9.85QGISLQSVYY134 pKa = 10.16IGRR137 pKa = 11.84GLLPAWPVQAKK148 pKa = 9.93QDD150 pKa = 3.13VDD152 pKa = 4.09LAIEE156 pKa = 4.37GAFEE160 pKa = 4.35VQDD163 pKa = 3.42EE164 pKa = 5.21SYY166 pKa = 11.67NSDD169 pKa = 3.21TDD171 pKa = 3.57GGSDD175 pKa = 3.45AEE177 pKa = 4.78GYY179 pKa = 7.94KK180 pKa = 9.91TDD182 pKa = 4.17GSDD185 pKa = 3.42EE186 pKa = 4.46TVTEE190 pKa = 4.32ATQLTPLAAQFKK202 pKa = 10.09QADD205 pKa = 4.3LSCSDD210 pKa = 4.43SDD212 pKa = 4.68GDD214 pKa = 5.24DD215 pKa = 3.68AGSDD219 pKa = 3.57DD220 pKa = 6.59DD221 pKa = 4.68YY222 pKa = 11.86DD223 pKa = 3.74QVIYY227 pKa = 10.25MNNFVCPEE235 pKa = 4.91PIYY238 pKa = 11.02EE239 pKa = 5.76DD240 pKa = 3.39IDD242 pKa = 3.98DD243 pKa = 4.0VNIAHH248 pKa = 6.32NVDD251 pKa = 3.99VVDD254 pKa = 4.3HH255 pKa = 6.11SQMVDD260 pKa = 3.03MQVFLVEE267 pKa = 4.41DD268 pKa = 4.5DD269 pKa = 3.76EE270 pKa = 7.19DD271 pKa = 3.84IAHH274 pKa = 6.24VPMAEE279 pKa = 4.12LAVDD283 pKa = 3.59YY284 pKa = 10.91DD285 pKa = 3.85GYY287 pKa = 11.38EE288 pKa = 4.34YY289 pKa = 11.5YY290 pKa = 10.8CGDD293 pKa = 4.15SGDD296 pKa = 3.72EE297 pKa = 4.67AIDD300 pKa = 3.97IEE302 pKa = 4.39LQDD305 pKa = 3.71GEE307 pKa = 4.99LYY309 pKa = 10.99CFVGRR314 pKa = 11.84EE315 pKa = 3.88KK316 pKa = 10.81SDD318 pKa = 3.49EE319 pKa = 4.09EE320 pKa = 4.65GEE322 pKa = 3.96EE323 pKa = 4.25DD324 pKa = 3.73EE325 pKa = 5.24QEE327 pKa = 4.18EE328 pKa = 4.52AMPMSDD334 pKa = 3.66GQLSSDD340 pKa = 3.88SQHH343 pKa = 7.2DD344 pKa = 3.87DD345 pKa = 3.5VGDD348 pKa = 3.76SDD350 pKa = 4.58EE351 pKa = 5.95DD352 pKa = 3.88GTATTTDD359 pKa = 3.96DD360 pKa = 4.92DD361 pKa = 4.7SSLSEE366 pKa = 4.06GSEE369 pKa = 3.89SDD371 pKa = 3.71TSIEE375 pKa = 4.2DD376 pKa = 3.21
MM1 pKa = 7.68SDD3 pKa = 3.05MAGEE7 pKa = 4.19LQPLGSQATSIAVDD21 pKa = 3.62EE22 pKa = 4.53FTKK25 pKa = 10.51FLKK28 pKa = 10.85NSVPTAMIPIDD39 pKa = 3.15IHH41 pKa = 5.91KK42 pKa = 10.36CLVYY46 pKa = 9.58TANCILDD53 pKa = 3.56VAKK56 pKa = 10.64EE57 pKa = 4.18SGVEE61 pKa = 3.8LSEE64 pKa = 3.69ITIGRR69 pKa = 11.84DD70 pKa = 4.05CMPTWPCCFWDD81 pKa = 3.78EE82 pKa = 4.5RR83 pKa = 11.84VEE85 pKa = 4.27TLHH88 pKa = 6.1EE89 pKa = 4.59TIQKK93 pKa = 10.13GLVVSQMVRR102 pKa = 11.84SLGDD106 pKa = 3.16FVRR109 pKa = 11.84LEE111 pKa = 4.48IINKK115 pKa = 9.73GNMKK119 pKa = 10.1LDD121 pKa = 3.7EE122 pKa = 4.43ARR124 pKa = 11.84KK125 pKa = 9.85QGISLQSVYY134 pKa = 10.16IGRR137 pKa = 11.84GLLPAWPVQAKK148 pKa = 9.93QDD150 pKa = 3.13VDD152 pKa = 4.09LAIEE156 pKa = 4.37GAFEE160 pKa = 4.35VQDD163 pKa = 3.42EE164 pKa = 5.21SYY166 pKa = 11.67NSDD169 pKa = 3.21TDD171 pKa = 3.57GGSDD175 pKa = 3.45AEE177 pKa = 4.78GYY179 pKa = 7.94KK180 pKa = 9.91TDD182 pKa = 4.17GSDD185 pKa = 3.42EE186 pKa = 4.46TVTEE190 pKa = 4.32ATQLTPLAAQFKK202 pKa = 10.09QADD205 pKa = 4.3LSCSDD210 pKa = 4.43SDD212 pKa = 4.68GDD214 pKa = 5.24DD215 pKa = 3.68AGSDD219 pKa = 3.57DD220 pKa = 6.59DD221 pKa = 4.68YY222 pKa = 11.86DD223 pKa = 3.74QVIYY227 pKa = 10.25MNNFVCPEE235 pKa = 4.91PIYY238 pKa = 11.02EE239 pKa = 5.76DD240 pKa = 3.39IDD242 pKa = 3.98DD243 pKa = 4.0VNIAHH248 pKa = 6.32NVDD251 pKa = 3.99VVDD254 pKa = 4.3HH255 pKa = 6.11SQMVDD260 pKa = 3.03MQVFLVEE267 pKa = 4.41DD268 pKa = 4.5DD269 pKa = 3.76EE270 pKa = 7.19DD271 pKa = 3.84IAHH274 pKa = 6.24VPMAEE279 pKa = 4.12LAVDD283 pKa = 3.59YY284 pKa = 10.91DD285 pKa = 3.85GYY287 pKa = 11.38EE288 pKa = 4.34YY289 pKa = 11.5YY290 pKa = 10.8CGDD293 pKa = 4.15SGDD296 pKa = 3.72EE297 pKa = 4.67AIDD300 pKa = 3.97IEE302 pKa = 4.39LQDD305 pKa = 3.71GEE307 pKa = 4.99LYY309 pKa = 10.99CFVGRR314 pKa = 11.84EE315 pKa = 3.88KK316 pKa = 10.81SDD318 pKa = 3.49EE319 pKa = 4.09EE320 pKa = 4.65GEE322 pKa = 3.96EE323 pKa = 4.25DD324 pKa = 3.73EE325 pKa = 5.24QEE327 pKa = 4.18EE328 pKa = 4.52AMPMSDD334 pKa = 3.66GQLSSDD340 pKa = 3.88SQHH343 pKa = 7.2DD344 pKa = 3.87DD345 pKa = 3.5VGDD348 pKa = 3.76SDD350 pKa = 4.58EE351 pKa = 5.95DD352 pKa = 3.88GTATTTDD359 pKa = 3.96DD360 pKa = 4.92DD361 pKa = 4.7SSLSEE366 pKa = 4.06GSEE369 pKa = 3.89SDD371 pKa = 3.71TSIEE375 pKa = 4.2DD376 pKa = 3.21
Molecular weight: 41.26 kDa
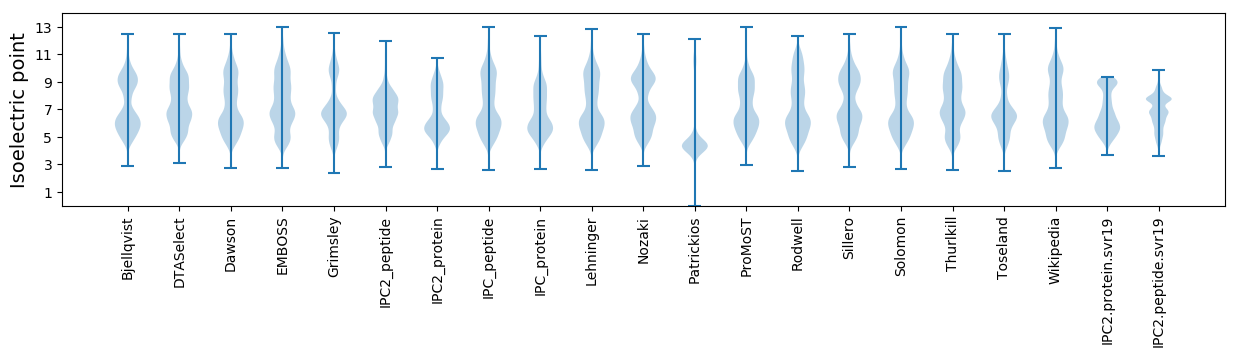
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C5XC56|A0A2C5XC56_9HYPO Uncharacterized protein OS=Ophiocordyceps australis OX=1399860 GN=CDD81_5251 PE=3 SV=1
MM1 pKa = 7.7AISARR6 pKa = 11.84RR7 pKa = 11.84VILPQWQWRR16 pKa = 11.84CVSSSTSSSQHH27 pKa = 5.87RR28 pKa = 11.84LPHH31 pKa = 6.29RR32 pKa = 11.84LHH34 pKa = 6.87RR35 pKa = 11.84PNPPRR40 pKa = 11.84KK41 pKa = 7.94HH42 pKa = 5.5TAPTSQPEE50 pKa = 4.31TLLSLWRR57 pKa = 11.84RR58 pKa = 11.84SWMPLTGAALAAGFLGFYY76 pKa = 10.38IVGTVAASLRR86 pKa = 11.84QPCQLPNTACRR97 pKa = 11.84TAIPTGRR104 pKa = 11.84PSALTGEE111 pKa = 4.34NVEE114 pKa = 4.36TFDD117 pKa = 6.42RR118 pKa = 11.84EE119 pKa = 4.19LDD121 pKa = 3.51WPEE124 pKa = 2.82WWMGISRR131 pKa = 11.84LRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84IGALAAGDD143 pKa = 3.87VLEE146 pKa = 4.44VAVGTGRR153 pKa = 11.84NLTFYY158 pKa = 9.31QWKK161 pKa = 8.0PASPARR167 pKa = 11.84KK168 pKa = 8.9NRR170 pKa = 11.84PRR172 pKa = 11.84GITSFTGLDD181 pKa = 3.1ISADD185 pKa = 3.76MLVLAQKK192 pKa = 10.74RR193 pKa = 11.84LDD195 pKa = 4.04LLHH198 pKa = 6.57TPATQFQSPTRR209 pKa = 11.84SSSAQNRR216 pKa = 11.84LRR218 pKa = 11.84LITANVQEE226 pKa = 5.06PLPPPPVTSVPVPGSTKK243 pKa = 10.05PCAKK247 pKa = 9.83YY248 pKa = 9.08DD249 pKa = 3.93TIIQTFGLCSVADD262 pKa = 3.63PVRR265 pKa = 11.84ALEE268 pKa = 4.06NLAAAVKK275 pKa = 9.27PGSGRR280 pKa = 11.84IILLEE285 pKa = 4.22HH286 pKa = 5.7GRR288 pKa = 11.84GWLGLVNGLLDD299 pKa = 3.94RR300 pKa = 11.84FAPGHH305 pKa = 4.86YY306 pKa = 9.68SKK308 pKa = 11.27YY309 pKa = 8.15GCWWNRR315 pKa = 11.84DD316 pKa = 3.13IEE318 pKa = 4.4GLIHH322 pKa = 5.74EE323 pKa = 6.17AIRR326 pKa = 11.84STPQLEE332 pKa = 4.1LVKK335 pKa = 10.2LHH337 pKa = 7.05RR338 pKa = 11.84PNIWQLGTIVWAEE351 pKa = 3.95LRR353 pKa = 11.84VALPPEE359 pKa = 4.34KK360 pKa = 10.73SS361 pKa = 3.09
MM1 pKa = 7.7AISARR6 pKa = 11.84RR7 pKa = 11.84VILPQWQWRR16 pKa = 11.84CVSSSTSSSQHH27 pKa = 5.87RR28 pKa = 11.84LPHH31 pKa = 6.29RR32 pKa = 11.84LHH34 pKa = 6.87RR35 pKa = 11.84PNPPRR40 pKa = 11.84KK41 pKa = 7.94HH42 pKa = 5.5TAPTSQPEE50 pKa = 4.31TLLSLWRR57 pKa = 11.84RR58 pKa = 11.84SWMPLTGAALAAGFLGFYY76 pKa = 10.38IVGTVAASLRR86 pKa = 11.84QPCQLPNTACRR97 pKa = 11.84TAIPTGRR104 pKa = 11.84PSALTGEE111 pKa = 4.34NVEE114 pKa = 4.36TFDD117 pKa = 6.42RR118 pKa = 11.84EE119 pKa = 4.19LDD121 pKa = 3.51WPEE124 pKa = 2.82WWMGISRR131 pKa = 11.84LRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84IGALAAGDD143 pKa = 3.87VLEE146 pKa = 4.44VAVGTGRR153 pKa = 11.84NLTFYY158 pKa = 9.31QWKK161 pKa = 8.0PASPARR167 pKa = 11.84KK168 pKa = 8.9NRR170 pKa = 11.84PRR172 pKa = 11.84GITSFTGLDD181 pKa = 3.1ISADD185 pKa = 3.76MLVLAQKK192 pKa = 10.74RR193 pKa = 11.84LDD195 pKa = 4.04LLHH198 pKa = 6.57TPATQFQSPTRR209 pKa = 11.84SSSAQNRR216 pKa = 11.84LRR218 pKa = 11.84LITANVQEE226 pKa = 5.06PLPPPPVTSVPVPGSTKK243 pKa = 10.05PCAKK247 pKa = 9.83YY248 pKa = 9.08DD249 pKa = 3.93TIIQTFGLCSVADD262 pKa = 3.63PVRR265 pKa = 11.84ALEE268 pKa = 4.06NLAAAVKK275 pKa = 9.27PGSGRR280 pKa = 11.84IILLEE285 pKa = 4.22HH286 pKa = 5.7GRR288 pKa = 11.84GWLGLVNGLLDD299 pKa = 3.94RR300 pKa = 11.84FAPGHH305 pKa = 4.86YY306 pKa = 9.68SKK308 pKa = 11.27YY309 pKa = 8.15GCWWNRR315 pKa = 11.84DD316 pKa = 3.13IEE318 pKa = 4.4GLIHH322 pKa = 5.74EE323 pKa = 6.17AIRR326 pKa = 11.84STPQLEE332 pKa = 4.1LVKK335 pKa = 10.2LHH337 pKa = 7.05RR338 pKa = 11.84PNIWQLGTIVWAEE351 pKa = 3.95LRR353 pKa = 11.84VALPPEE359 pKa = 4.34KK360 pKa = 10.73SS361 pKa = 3.09
Molecular weight: 40.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4004734 |
66 |
5810 |
490.3 |
54.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.788 ± 0.028 | 1.397 ± 0.011 |
5.868 ± 0.02 | 5.707 ± 0.029 |
3.394 ± 0.017 | 6.798 ± 0.026 |
2.64 ± 0.012 | 4.223 ± 0.021 |
4.542 ± 0.027 | 9.173 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.01 | 3.242 ± 0.014 |
5.997 ± 0.032 | 4.492 ± 0.023 |
6.595 ± 0.024 | 8.291 ± 0.035 |
5.425 ± 0.019 | 6.162 ± 0.024 |
1.43 ± 0.01 | 2.465 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
