
Glaciihabitans arcticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Glaciihabitans
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
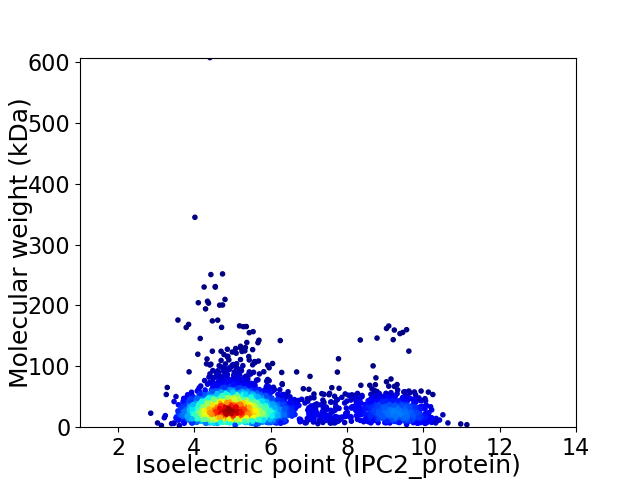
Virtual 2D-PAGE plot for 2984 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q9GXY9|A0A4Q9GXY9_9MICO ABC transporter substrate-binding protein OS=Glaciihabitans arcticus OX=2668039 GN=EYE40_12340 PE=3 SV=1
MM1 pKa = 7.81AEE3 pKa = 4.39GDD5 pKa = 4.35NPFPWGLTPGGKK17 pKa = 9.27PEE19 pKa = 5.1DD20 pKa = 4.46EE21 pKa = 4.55EE22 pKa = 4.76TPPTPPVAPEE32 pKa = 5.18PIDD35 pKa = 3.18QGYY38 pKa = 8.09FAAVTIEE45 pKa = 4.58EE46 pKa = 4.51PTSPAPEE53 pKa = 4.31GLPTEE58 pKa = 4.88AMASQEE64 pKa = 3.75WDD66 pKa = 3.68GPATEE71 pKa = 5.35AMPAQEE77 pKa = 4.26WEE79 pKa = 4.7GPPTDD84 pKa = 4.45PFPYY88 pKa = 9.5HH89 pKa = 6.59QPAPWEE95 pKa = 4.15PPPVDD100 pKa = 3.49EE101 pKa = 5.38SLGGTSNALAAQPIGLDD118 pKa = 3.39APVGEE123 pKa = 4.76SVPGEE128 pKa = 3.95NTDD131 pKa = 4.51AIEE134 pKa = 4.33DD135 pKa = 3.92LFGEE139 pKa = 5.04GSFQEE144 pKa = 4.36YY145 pKa = 9.77EE146 pKa = 3.91DD147 pKa = 5.22AIVVLEE153 pKa = 4.73GIPFPGKK160 pKa = 10.2AVEE163 pKa = 4.06PAAAPGALVAPPPTLLPAEE182 pKa = 4.27PMPRR186 pKa = 11.84LQRR189 pKa = 11.84NLLWAAGGLVAALALVAVFLGGVRR213 pKa = 11.84LAPQFAAAPEE223 pKa = 4.37APAVPSSAPTLAPEE237 pKa = 4.43SLGPLPAGVHH247 pKa = 5.46AWSDD251 pKa = 3.68LLGTEE256 pKa = 4.2CVEE259 pKa = 4.73PYY261 pKa = 10.39VSPWDD266 pKa = 3.61KK267 pKa = 10.93EE268 pKa = 4.23FTVVDD273 pKa = 3.63CSGPHH278 pKa = 5.36TAQLVATGVFEE289 pKa = 6.2DD290 pKa = 5.07SILDD294 pKa = 4.0AYY296 pKa = 9.86PGLEE300 pKa = 3.94EE301 pKa = 4.08LQSRR305 pKa = 11.84MGQLCSNATNVDD317 pKa = 3.28YY318 pKa = 9.2TAAKK322 pKa = 9.72AYY324 pKa = 11.0SDD326 pKa = 3.57IQVSASYY333 pKa = 10.62AADD336 pKa = 3.57EE337 pKa = 4.26QSWIDD342 pKa = 3.09GDD344 pKa = 4.0RR345 pKa = 11.84NYY347 pKa = 10.82FCFVSRR353 pKa = 11.84STGEE357 pKa = 3.98PLTSSVAMPDD367 pKa = 2.93RR368 pKa = 11.84PTPVVPVVPEE378 pKa = 4.0PEE380 pKa = 3.86PP381 pKa = 3.96
MM1 pKa = 7.81AEE3 pKa = 4.39GDD5 pKa = 4.35NPFPWGLTPGGKK17 pKa = 9.27PEE19 pKa = 5.1DD20 pKa = 4.46EE21 pKa = 4.55EE22 pKa = 4.76TPPTPPVAPEE32 pKa = 5.18PIDD35 pKa = 3.18QGYY38 pKa = 8.09FAAVTIEE45 pKa = 4.58EE46 pKa = 4.51PTSPAPEE53 pKa = 4.31GLPTEE58 pKa = 4.88AMASQEE64 pKa = 3.75WDD66 pKa = 3.68GPATEE71 pKa = 5.35AMPAQEE77 pKa = 4.26WEE79 pKa = 4.7GPPTDD84 pKa = 4.45PFPYY88 pKa = 9.5HH89 pKa = 6.59QPAPWEE95 pKa = 4.15PPPVDD100 pKa = 3.49EE101 pKa = 5.38SLGGTSNALAAQPIGLDD118 pKa = 3.39APVGEE123 pKa = 4.76SVPGEE128 pKa = 3.95NTDD131 pKa = 4.51AIEE134 pKa = 4.33DD135 pKa = 3.92LFGEE139 pKa = 5.04GSFQEE144 pKa = 4.36YY145 pKa = 9.77EE146 pKa = 3.91DD147 pKa = 5.22AIVVLEE153 pKa = 4.73GIPFPGKK160 pKa = 10.2AVEE163 pKa = 4.06PAAAPGALVAPPPTLLPAEE182 pKa = 4.27PMPRR186 pKa = 11.84LQRR189 pKa = 11.84NLLWAAGGLVAALALVAVFLGGVRR213 pKa = 11.84LAPQFAAAPEE223 pKa = 4.37APAVPSSAPTLAPEE237 pKa = 4.43SLGPLPAGVHH247 pKa = 5.46AWSDD251 pKa = 3.68LLGTEE256 pKa = 4.2CVEE259 pKa = 4.73PYY261 pKa = 10.39VSPWDD266 pKa = 3.61KK267 pKa = 10.93EE268 pKa = 4.23FTVVDD273 pKa = 3.63CSGPHH278 pKa = 5.36TAQLVATGVFEE289 pKa = 6.2DD290 pKa = 5.07SILDD294 pKa = 4.0AYY296 pKa = 9.86PGLEE300 pKa = 3.94EE301 pKa = 4.08LQSRR305 pKa = 11.84MGQLCSNATNVDD317 pKa = 3.28YY318 pKa = 9.2TAAKK322 pKa = 9.72AYY324 pKa = 11.0SDD326 pKa = 3.57IQVSASYY333 pKa = 10.62AADD336 pKa = 3.57EE337 pKa = 4.26QSWIDD342 pKa = 3.09GDD344 pKa = 4.0RR345 pKa = 11.84NYY347 pKa = 10.82FCFVSRR353 pKa = 11.84STGEE357 pKa = 3.98PLTSSVAMPDD367 pKa = 2.93RR368 pKa = 11.84PTPVVPVVPEE378 pKa = 4.0PEE380 pKa = 3.86PP381 pKa = 3.96
Molecular weight: 39.82 kDa
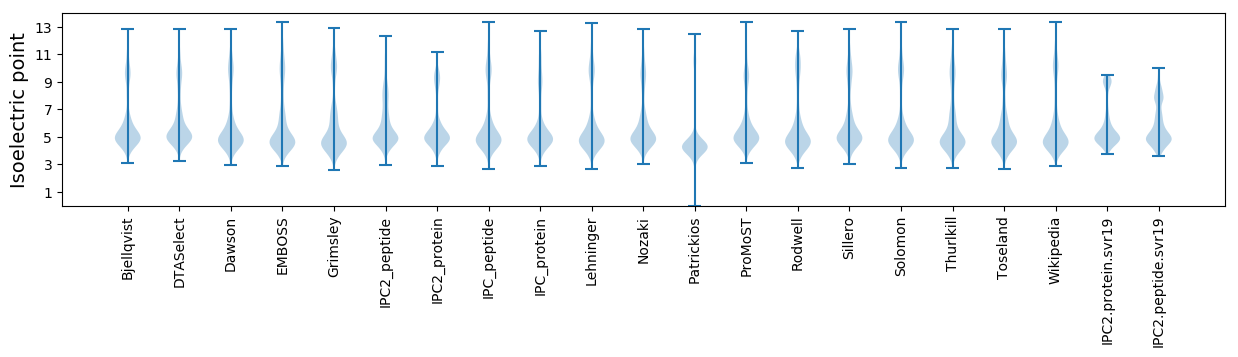
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q9GUL4|A0A4Q9GUL4_9MICO ADP-dependent (S)-NAD(P)H-hydrate dehydratase OS=Glaciihabitans arcticus OX=2668039 GN=nnrD PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
981908 |
24 |
6078 |
329.1 |
35.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.682 ± 0.054 | 0.447 ± 0.011 |
5.941 ± 0.041 | 5.608 ± 0.057 |
3.377 ± 0.03 | 8.856 ± 0.053 |
1.748 ± 0.024 | 4.98 ± 0.036 |
2.422 ± 0.037 | 10.316 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.652 ± 0.02 | 2.393 ± 0.037 |
5.191 ± 0.035 | 2.71 ± 0.023 |
6.311 ± 0.062 | 6.315 ± 0.041 |
6.578 ± 0.089 | 8.909 ± 0.041 |
1.465 ± 0.018 | 2.097 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
