
Beet soil-borne mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Hepelivirales; Benyviridae; Benyvirus
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
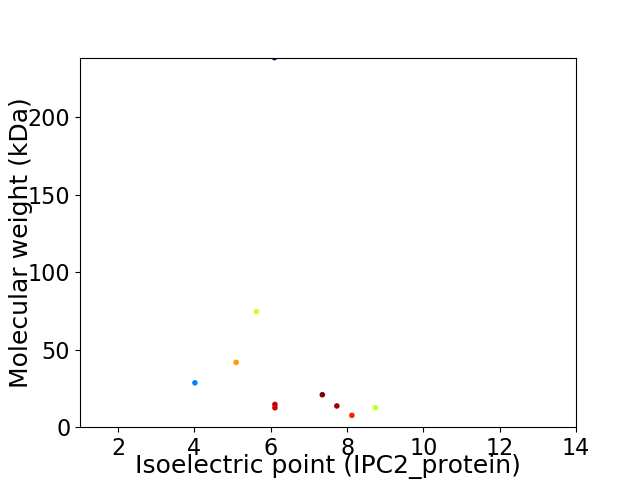
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9IF44|Q9IF44_9VIRU RNA-dependent RNA polymerase OS=Beet soil-borne mosaic virus OX=76343 PE=4 SV=1
MM1 pKa = 8.03DD2 pKa = 6.43LNTMMPAFNVAYY14 pKa = 8.8WDD16 pKa = 4.22GVHH19 pKa = 6.58APYY22 pKa = 10.25VVKK25 pKa = 10.73RR26 pKa = 11.84MMHH29 pKa = 5.02EE30 pKa = 3.87VVMNVGPAGFICYY43 pKa = 7.92PLPVDD48 pKa = 5.47FDD50 pKa = 5.11LNDD53 pKa = 3.33TGVIHH58 pKa = 6.77NFAYY62 pKa = 10.25HH63 pKa = 5.85NRR65 pKa = 11.84VKK67 pKa = 10.24TMRR70 pKa = 11.84LFVGIQNNCSEE81 pKa = 4.35WVYY84 pKa = 10.85GRR86 pKa = 11.84ARR88 pKa = 11.84FVVFSTSAISPWVNNGCMSLFSPFVGVNSPIDD120 pKa = 3.65RR121 pKa = 11.84NLLRR125 pKa = 11.84RR126 pKa = 11.84DD127 pKa = 3.25SRR129 pKa = 11.84GVSVLWDD136 pKa = 3.03RR137 pKa = 11.84VYY139 pKa = 10.79RR140 pKa = 11.84VNRR143 pKa = 11.84GTQLFVDD150 pKa = 3.84EE151 pKa = 4.82TFNFIGPGNYY161 pKa = 7.85PAQVGEE167 pKa = 4.63NYY169 pKa = 10.16PSATTYY175 pKa = 11.26DD176 pKa = 4.26SIYY179 pKa = 9.46VACVTDD185 pKa = 4.52WIDD188 pKa = 3.54NNVFRR193 pKa = 11.84LTSDD197 pKa = 3.2SVGWFHH203 pKa = 7.59SGLDD207 pKa = 3.67DD208 pKa = 4.6GPRR211 pKa = 11.84LAFGQGLNAPDD222 pKa = 4.79DD223 pKa = 4.92DD224 pKa = 5.77GDD226 pKa = 4.24GVVGDD231 pKa = 5.23DD232 pKa = 4.86DD233 pKa = 5.07VDD235 pKa = 3.72VDD237 pKa = 4.9GEE239 pKa = 4.56NIDD242 pKa = 3.83EE243 pKa = 5.05DD244 pKa = 5.11ADD246 pKa = 4.05VMDD249 pKa = 5.99DD250 pKa = 4.46ANTDD254 pKa = 3.52DD255 pKa = 5.05GDD257 pKa = 3.59
MM1 pKa = 8.03DD2 pKa = 6.43LNTMMPAFNVAYY14 pKa = 8.8WDD16 pKa = 4.22GVHH19 pKa = 6.58APYY22 pKa = 10.25VVKK25 pKa = 10.73RR26 pKa = 11.84MMHH29 pKa = 5.02EE30 pKa = 3.87VVMNVGPAGFICYY43 pKa = 7.92PLPVDD48 pKa = 5.47FDD50 pKa = 5.11LNDD53 pKa = 3.33TGVIHH58 pKa = 6.77NFAYY62 pKa = 10.25HH63 pKa = 5.85NRR65 pKa = 11.84VKK67 pKa = 10.24TMRR70 pKa = 11.84LFVGIQNNCSEE81 pKa = 4.35WVYY84 pKa = 10.85GRR86 pKa = 11.84ARR88 pKa = 11.84FVVFSTSAISPWVNNGCMSLFSPFVGVNSPIDD120 pKa = 3.65RR121 pKa = 11.84NLLRR125 pKa = 11.84RR126 pKa = 11.84DD127 pKa = 3.25SRR129 pKa = 11.84GVSVLWDD136 pKa = 3.03RR137 pKa = 11.84VYY139 pKa = 10.79RR140 pKa = 11.84VNRR143 pKa = 11.84GTQLFVDD150 pKa = 3.84EE151 pKa = 4.82TFNFIGPGNYY161 pKa = 7.85PAQVGEE167 pKa = 4.63NYY169 pKa = 10.16PSATTYY175 pKa = 11.26DD176 pKa = 4.26SIYY179 pKa = 9.46VACVTDD185 pKa = 4.52WIDD188 pKa = 3.54NNVFRR193 pKa = 11.84LTSDD197 pKa = 3.2SVGWFHH203 pKa = 7.59SGLDD207 pKa = 3.67DD208 pKa = 4.6GPRR211 pKa = 11.84LAFGQGLNAPDD222 pKa = 4.79DD223 pKa = 4.92DD224 pKa = 5.77GDD226 pKa = 4.24GVVGDD231 pKa = 5.23DD232 pKa = 4.86DD233 pKa = 5.07VDD235 pKa = 3.72VDD237 pKa = 4.9GEE239 pKa = 4.56NIDD242 pKa = 3.83EE243 pKa = 5.05DD244 pKa = 5.11ADD246 pKa = 4.05VMDD249 pKa = 5.99DD250 pKa = 4.46ANTDD254 pKa = 3.52DD255 pKa = 5.05GDD257 pKa = 3.59
Molecular weight: 28.63 kDa
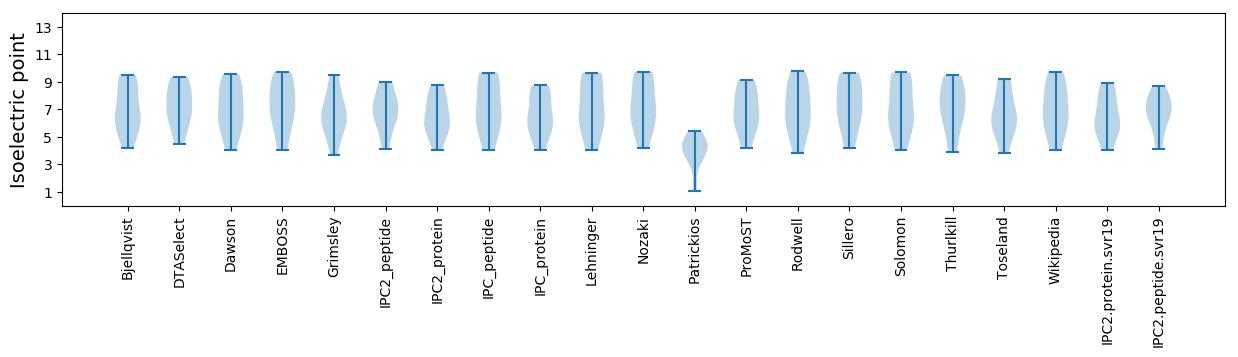
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O72593|O72593_9VIRU 8kDa protein OS=Beet soil-borne mosaic virus OX=76343 PE=4 SV=1
MM1 pKa = 7.3SRR3 pKa = 11.84EE4 pKa = 3.36ITARR8 pKa = 11.84ANKK11 pKa = 9.43NVPIVVGVCVVAFFVLLAFMQQKK34 pKa = 10.37HH35 pKa = 5.0KK36 pKa = 8.76THH38 pKa = 6.44SGGDD42 pKa = 3.43YY43 pKa = 10.74GVPTFSNGGKK53 pKa = 9.9YY54 pKa = 9.73RR55 pKa = 11.84DD56 pKa = 3.9GTRR59 pKa = 11.84SADD62 pKa = 3.47FNSNNHH68 pKa = 5.99RR69 pKa = 11.84AYY71 pKa = 10.32GCGGSKK77 pKa = 10.41SSVTGKK83 pKa = 10.29VGQQLLVLALVVAVFVLFMRR103 pKa = 11.84GCWSSPEE110 pKa = 4.39HH111 pKa = 6.31ICNGSCGG118 pKa = 3.53
MM1 pKa = 7.3SRR3 pKa = 11.84EE4 pKa = 3.36ITARR8 pKa = 11.84ANKK11 pKa = 9.43NVPIVVGVCVVAFFVLLAFMQQKK34 pKa = 10.37HH35 pKa = 5.0KK36 pKa = 8.76THH38 pKa = 6.44SGGDD42 pKa = 3.43YY43 pKa = 10.74GVPTFSNGGKK53 pKa = 9.9YY54 pKa = 9.73RR55 pKa = 11.84DD56 pKa = 3.9GTRR59 pKa = 11.84SADD62 pKa = 3.47FNSNNHH68 pKa = 5.99RR69 pKa = 11.84AYY71 pKa = 10.32GCGGSKK77 pKa = 10.41SSVTGKK83 pKa = 10.29VGQQLLVLALVVAVFVLFMRR103 pKa = 11.84GCWSSPEE110 pKa = 4.39HH111 pKa = 6.31ICNGSCGG118 pKa = 3.53
Molecular weight: 12.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4181 |
68 |
2117 |
418.1 |
46.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.175 ± 0.601 | 1.961 ± 0.471 |
6.649 ± 0.777 | 4.855 ± 0.532 |
4.831 ± 0.35 | 6.817 ± 0.698 |
2.009 ± 0.2 | 4.712 ± 0.433 |
5.142 ± 0.786 | 9.137 ± 0.774 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.631 ± 0.226 | 4.951 ± 0.429 |
3.803 ± 0.335 | 2.583 ± 0.368 |
5.429 ± 0.35 | 7.917 ± 0.286 |
5.214 ± 0.545 | 9.352 ± 0.812 |
1.722 ± 0.203 | 3.109 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
