
Rhodobacteraceae bacterium PD-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
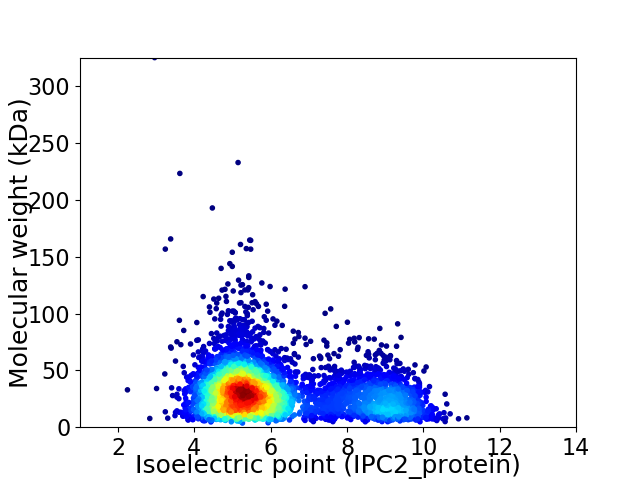
Virtual 2D-PAGE plot for 4616 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
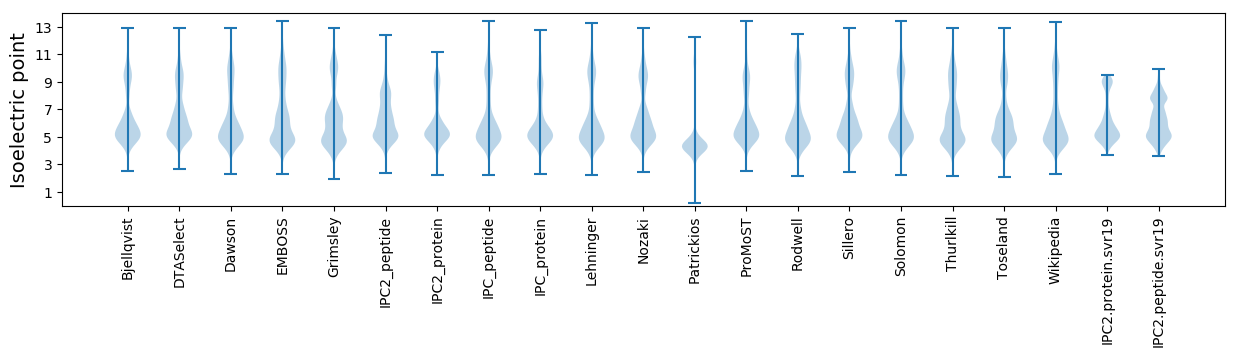
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4ERK4|A0A0B4ERK4_9RHOB 3-isopropylmalate dehydratase (Fragment) OS=Rhodobacteraceae bacterium PD-2 OX=1169855 GN=P279_30875 PE=3 SV=1
MM1 pKa = 7.16ATYY4 pKa = 10.36VYY6 pKa = 10.85DD7 pKa = 3.75GTSLNNLILDD17 pKa = 4.12PASGFDD23 pKa = 3.48VYY25 pKa = 11.54DD26 pKa = 3.97DD27 pKa = 4.15SFYY30 pKa = 11.43SSTLGDD36 pKa = 3.44RR37 pKa = 11.84FDD39 pKa = 4.41NLFDD43 pKa = 3.66LTGITAYY50 pKa = 10.75VDD52 pKa = 3.38GSIFEE57 pKa = 4.25MLAGNDD63 pKa = 3.35QFLGSEE69 pKa = 3.88LGEE72 pKa = 3.96NVFGGEE78 pKa = 4.55GNDD81 pKa = 3.43SLTGNGGNDD90 pKa = 3.89SLWGGLGADD99 pKa = 3.9TLRR102 pKa = 11.84GGSGNDD108 pKa = 3.28QLVGDD113 pKa = 4.33QGVDD117 pKa = 3.37LLDD120 pKa = 4.17GGEE123 pKa = 4.25GNDD126 pKa = 3.71LFLFGSGGDD135 pKa = 3.56VDD137 pKa = 3.82ILRR140 pKa = 11.84GGSGTDD146 pKa = 2.53IGYY149 pKa = 10.77YY150 pKa = 10.04SGDD153 pKa = 3.64YY154 pKa = 10.51LQGLIVDD161 pKa = 4.48AASSIEE167 pKa = 3.92VLHH170 pKa = 5.72QNSYY174 pKa = 9.83NGYY177 pKa = 9.36YY178 pKa = 10.37GDD180 pKa = 3.64NAANLFDD187 pKa = 3.72VSGVQSYY194 pKa = 10.71VGAADD199 pKa = 5.01FYY201 pKa = 11.45LGGGNDD207 pKa = 3.54TFVGSDD213 pKa = 3.25VAEE216 pKa = 3.94WAMGQFGNDD225 pKa = 3.14TLLGNGGDD233 pKa = 4.55DD234 pKa = 4.76RR235 pKa = 11.84LQGWEE240 pKa = 4.87DD241 pKa = 4.01DD242 pKa = 3.75DD243 pKa = 4.93LLRR246 pKa = 11.84GGAGADD252 pKa = 3.52TLEE255 pKa = 4.91GGSGTDD261 pKa = 3.07TADD264 pKa = 3.51YY265 pKa = 9.14TGSSSWVNVSLLTGYY280 pKa = 9.18TSGGHH285 pKa = 5.43AQGDD289 pKa = 3.97VLSGIEE295 pKa = 4.1NLTGSAHH302 pKa = 6.87GDD304 pKa = 3.39RR305 pKa = 11.84LSGDD309 pKa = 2.9HH310 pKa = 6.47GANVLRR316 pKa = 11.84GQAGDD321 pKa = 4.01DD322 pKa = 3.29ILRR325 pKa = 11.84GRR327 pKa = 11.84GGADD331 pKa = 3.05TLEE334 pKa = 4.67GGAGSDD340 pKa = 2.85IADD343 pKa = 3.74YY344 pKa = 11.01SEE346 pKa = 4.06SAGWVNVSLISGYY359 pKa = 11.01AGGGAGSHH367 pKa = 7.15AIGDD371 pKa = 3.54VWIGIEE377 pKa = 4.05NLIGSDD383 pKa = 3.67FADD386 pKa = 4.52RR387 pKa = 11.84LNGDD391 pKa = 3.06NGEE394 pKa = 4.21NVLEE398 pKa = 4.42GRR400 pKa = 11.84AGGDD404 pKa = 2.7IFNGYY409 pKa = 9.22GGSDD413 pKa = 3.4TLSYY417 pKa = 10.56ASSVAWVNVSLLTGYY432 pKa = 8.81TGGGAGNHH440 pKa = 6.41AAGDD444 pKa = 3.57SWTSIEE450 pKa = 4.03NLRR453 pKa = 11.84GSAHH457 pKa = 7.32ADD459 pKa = 3.47RR460 pKa = 11.84LNGDD464 pKa = 3.1HH465 pKa = 6.82GANVLEE471 pKa = 5.01GGAGGDD477 pKa = 3.07IVNGNGGSDD486 pKa = 3.43TLSYY490 pKa = 10.89ASSEE494 pKa = 3.7AWVNVSLLTGYY505 pKa = 8.81TGGGAGNHH513 pKa = 6.41AAGDD517 pKa = 3.57SWTSIEE523 pKa = 4.03NLRR526 pKa = 11.84GSAHH530 pKa = 7.32ADD532 pKa = 3.47RR533 pKa = 11.84LNGDD537 pKa = 3.1HH538 pKa = 6.82GANVLEE544 pKa = 5.01GGAGGDD550 pKa = 3.07IVNGNGGSDD559 pKa = 3.43TLSYY563 pKa = 10.89ASSEE567 pKa = 3.7AWVNVSLLTGYY578 pKa = 8.81TGGGAGNHH586 pKa = 6.41AAGDD590 pKa = 3.57SWTSIEE596 pKa = 4.03NLRR599 pKa = 11.84GSAHH603 pKa = 7.32ADD605 pKa = 3.47RR606 pKa = 11.84LNGDD610 pKa = 3.1HH611 pKa = 6.82GANVLEE617 pKa = 5.01GGAGGDD623 pKa = 3.07IVNGNGGSDD632 pKa = 3.43TLSYY636 pKa = 10.89ASSEE640 pKa = 3.7AWVNVSLLTGYY651 pKa = 8.81TGGGAGNHH659 pKa = 6.41AAGDD663 pKa = 3.57SWTSIEE669 pKa = 4.03NLRR672 pKa = 11.84GSAHH676 pKa = 7.32ADD678 pKa = 3.47RR679 pKa = 11.84LNGDD683 pKa = 3.1HH684 pKa = 6.82GANVLEE690 pKa = 5.01GGAGGDD696 pKa = 3.07IVNGNGGSDD705 pKa = 3.43TLSYY709 pKa = 10.56ASSVAWVNVSLLTGYY724 pKa = 8.81TGGGAGNHH732 pKa = 6.41AAGDD736 pKa = 3.57SWTSIEE742 pKa = 4.03NLRR745 pKa = 11.84GSAHH749 pKa = 7.32ADD751 pKa = 3.47RR752 pKa = 11.84LNGDD756 pKa = 3.1HH757 pKa = 6.82GANVLEE763 pKa = 4.91GGAGNDD769 pKa = 3.04ILRR772 pKa = 11.84GNGGIDD778 pKa = 3.04TFVFADD784 pKa = 4.05GFGSDD789 pKa = 5.02SVLDD793 pKa = 3.84YY794 pKa = 11.64QNGSEE799 pKa = 4.84KK800 pKa = 10.58FDD802 pKa = 3.8FTDD805 pKa = 3.11HH806 pKa = 6.82AGVSGFGDD814 pKa = 3.7LTVSASGADD823 pKa = 3.55ALIADD828 pKa = 4.68GFGNQITVTGAAGLIDD844 pKa = 3.67ATDD847 pKa = 4.35FIFF850 pKa = 5.29
MM1 pKa = 7.16ATYY4 pKa = 10.36VYY6 pKa = 10.85DD7 pKa = 3.75GTSLNNLILDD17 pKa = 4.12PASGFDD23 pKa = 3.48VYY25 pKa = 11.54DD26 pKa = 3.97DD27 pKa = 4.15SFYY30 pKa = 11.43SSTLGDD36 pKa = 3.44RR37 pKa = 11.84FDD39 pKa = 4.41NLFDD43 pKa = 3.66LTGITAYY50 pKa = 10.75VDD52 pKa = 3.38GSIFEE57 pKa = 4.25MLAGNDD63 pKa = 3.35QFLGSEE69 pKa = 3.88LGEE72 pKa = 3.96NVFGGEE78 pKa = 4.55GNDD81 pKa = 3.43SLTGNGGNDD90 pKa = 3.89SLWGGLGADD99 pKa = 3.9TLRR102 pKa = 11.84GGSGNDD108 pKa = 3.28QLVGDD113 pKa = 4.33QGVDD117 pKa = 3.37LLDD120 pKa = 4.17GGEE123 pKa = 4.25GNDD126 pKa = 3.71LFLFGSGGDD135 pKa = 3.56VDD137 pKa = 3.82ILRR140 pKa = 11.84GGSGTDD146 pKa = 2.53IGYY149 pKa = 10.77YY150 pKa = 10.04SGDD153 pKa = 3.64YY154 pKa = 10.51LQGLIVDD161 pKa = 4.48AASSIEE167 pKa = 3.92VLHH170 pKa = 5.72QNSYY174 pKa = 9.83NGYY177 pKa = 9.36YY178 pKa = 10.37GDD180 pKa = 3.64NAANLFDD187 pKa = 3.72VSGVQSYY194 pKa = 10.71VGAADD199 pKa = 5.01FYY201 pKa = 11.45LGGGNDD207 pKa = 3.54TFVGSDD213 pKa = 3.25VAEE216 pKa = 3.94WAMGQFGNDD225 pKa = 3.14TLLGNGGDD233 pKa = 4.55DD234 pKa = 4.76RR235 pKa = 11.84LQGWEE240 pKa = 4.87DD241 pKa = 4.01DD242 pKa = 3.75DD243 pKa = 4.93LLRR246 pKa = 11.84GGAGADD252 pKa = 3.52TLEE255 pKa = 4.91GGSGTDD261 pKa = 3.07TADD264 pKa = 3.51YY265 pKa = 9.14TGSSSWVNVSLLTGYY280 pKa = 9.18TSGGHH285 pKa = 5.43AQGDD289 pKa = 3.97VLSGIEE295 pKa = 4.1NLTGSAHH302 pKa = 6.87GDD304 pKa = 3.39RR305 pKa = 11.84LSGDD309 pKa = 2.9HH310 pKa = 6.47GANVLRR316 pKa = 11.84GQAGDD321 pKa = 4.01DD322 pKa = 3.29ILRR325 pKa = 11.84GRR327 pKa = 11.84GGADD331 pKa = 3.05TLEE334 pKa = 4.67GGAGSDD340 pKa = 2.85IADD343 pKa = 3.74YY344 pKa = 11.01SEE346 pKa = 4.06SAGWVNVSLISGYY359 pKa = 11.01AGGGAGSHH367 pKa = 7.15AIGDD371 pKa = 3.54VWIGIEE377 pKa = 4.05NLIGSDD383 pKa = 3.67FADD386 pKa = 4.52RR387 pKa = 11.84LNGDD391 pKa = 3.06NGEE394 pKa = 4.21NVLEE398 pKa = 4.42GRR400 pKa = 11.84AGGDD404 pKa = 2.7IFNGYY409 pKa = 9.22GGSDD413 pKa = 3.4TLSYY417 pKa = 10.56ASSVAWVNVSLLTGYY432 pKa = 8.81TGGGAGNHH440 pKa = 6.41AAGDD444 pKa = 3.57SWTSIEE450 pKa = 4.03NLRR453 pKa = 11.84GSAHH457 pKa = 7.32ADD459 pKa = 3.47RR460 pKa = 11.84LNGDD464 pKa = 3.1HH465 pKa = 6.82GANVLEE471 pKa = 5.01GGAGGDD477 pKa = 3.07IVNGNGGSDD486 pKa = 3.43TLSYY490 pKa = 10.89ASSEE494 pKa = 3.7AWVNVSLLTGYY505 pKa = 8.81TGGGAGNHH513 pKa = 6.41AAGDD517 pKa = 3.57SWTSIEE523 pKa = 4.03NLRR526 pKa = 11.84GSAHH530 pKa = 7.32ADD532 pKa = 3.47RR533 pKa = 11.84LNGDD537 pKa = 3.1HH538 pKa = 6.82GANVLEE544 pKa = 5.01GGAGGDD550 pKa = 3.07IVNGNGGSDD559 pKa = 3.43TLSYY563 pKa = 10.89ASSEE567 pKa = 3.7AWVNVSLLTGYY578 pKa = 8.81TGGGAGNHH586 pKa = 6.41AAGDD590 pKa = 3.57SWTSIEE596 pKa = 4.03NLRR599 pKa = 11.84GSAHH603 pKa = 7.32ADD605 pKa = 3.47RR606 pKa = 11.84LNGDD610 pKa = 3.1HH611 pKa = 6.82GANVLEE617 pKa = 5.01GGAGGDD623 pKa = 3.07IVNGNGGSDD632 pKa = 3.43TLSYY636 pKa = 10.89ASSEE640 pKa = 3.7AWVNVSLLTGYY651 pKa = 8.81TGGGAGNHH659 pKa = 6.41AAGDD663 pKa = 3.57SWTSIEE669 pKa = 4.03NLRR672 pKa = 11.84GSAHH676 pKa = 7.32ADD678 pKa = 3.47RR679 pKa = 11.84LNGDD683 pKa = 3.1HH684 pKa = 6.82GANVLEE690 pKa = 5.01GGAGGDD696 pKa = 3.07IVNGNGGSDD705 pKa = 3.43TLSYY709 pKa = 10.56ASSVAWVNVSLLTGYY724 pKa = 8.81TGGGAGNHH732 pKa = 6.41AAGDD736 pKa = 3.57SWTSIEE742 pKa = 4.03NLRR745 pKa = 11.84GSAHH749 pKa = 7.32ADD751 pKa = 3.47RR752 pKa = 11.84LNGDD756 pKa = 3.1HH757 pKa = 6.82GANVLEE763 pKa = 4.91GGAGNDD769 pKa = 3.04ILRR772 pKa = 11.84GNGGIDD778 pKa = 3.04TFVFADD784 pKa = 4.05GFGSDD789 pKa = 5.02SVLDD793 pKa = 3.84YY794 pKa = 11.64QNGSEE799 pKa = 4.84KK800 pKa = 10.58FDD802 pKa = 3.8FTDD805 pKa = 3.11HH806 pKa = 6.82AGVSGFGDD814 pKa = 3.7LTVSASGADD823 pKa = 3.55ALIADD828 pKa = 4.68GFGNQITVTGAAGLIDD844 pKa = 3.67ATDD847 pKa = 4.35FIFF850 pKa = 5.29
Molecular weight: 85.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4FJ82|A0A0B4FJ82_9RHOB Flagellar basal body rod protein FlgB (Fragment) OS=Rhodobacteraceae bacterium PD-2 OX=1169855 GN=flgB PE=3 SV=1
MM1 pKa = 7.45NSATIFNFRR10 pKa = 11.84RR11 pKa = 11.84GLPAVFHH18 pKa = 5.55VRR20 pKa = 11.84RR21 pKa = 11.84FIIVGPAHH29 pKa = 7.03LGGFLLRR36 pKa = 11.84TPMHH40 pKa = 6.78GPQIHH45 pKa = 5.47VLKK48 pKa = 10.59AALPLWHH55 pKa = 6.9MSSKK59 pKa = 10.01HH60 pKa = 5.13RR61 pKa = 11.84HH62 pKa = 4.43RR63 pKa = 11.84QAPRR67 pKa = 11.84RR68 pKa = 11.84VSAQQ72 pKa = 2.96
MM1 pKa = 7.45NSATIFNFRR10 pKa = 11.84RR11 pKa = 11.84GLPAVFHH18 pKa = 5.55VRR20 pKa = 11.84RR21 pKa = 11.84FIIVGPAHH29 pKa = 7.03LGGFLLRR36 pKa = 11.84TPMHH40 pKa = 6.78GPQIHH45 pKa = 5.47VLKK48 pKa = 10.59AALPLWHH55 pKa = 6.9MSSKK59 pKa = 10.01HH60 pKa = 5.13RR61 pKa = 11.84HH62 pKa = 4.43RR63 pKa = 11.84QAPRR67 pKa = 11.84RR68 pKa = 11.84VSAQQ72 pKa = 2.96
Molecular weight: 8.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1370329 |
34 |
3204 |
296.9 |
32.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.64 ± 0.052 | 0.917 ± 0.011 |
5.979 ± 0.029 | 5.986 ± 0.036 |
3.624 ± 0.024 | 8.955 ± 0.036 |
2.054 ± 0.018 | 4.806 ± 0.031 |
2.844 ± 0.029 | 10.233 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.812 ± 0.018 | 2.38 ± 0.02 |
5.186 ± 0.027 | 3.136 ± 0.021 |
6.986 ± 0.044 | 4.937 ± 0.028 |
5.523 ± 0.037 | 7.428 ± 0.032 |
1.4 ± 0.016 | 2.175 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
