
Sanxia narna-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.89
Get precalculated fractions of proteins
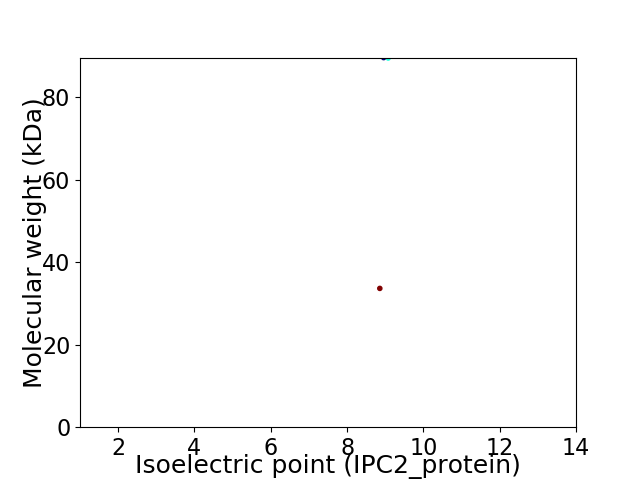
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIL2|A0A1L3KIL2_9VIRU RNA-dependent RNA polymerase OS=Sanxia narna-like virus 1 OX=1923360 PE=4 SV=1
MM1 pKa = 7.87KK2 pKa = 10.2YY3 pKa = 11.09NLMTKK8 pKa = 10.1SKK10 pKa = 9.14TKK12 pKa = 10.66SNQKK16 pKa = 8.64PAQRR20 pKa = 11.84AKK22 pKa = 10.31PRR24 pKa = 11.84RR25 pKa = 11.84SRR27 pKa = 11.84QRR29 pKa = 11.84ANKK32 pKa = 8.91VQDD35 pKa = 3.52GLDD38 pKa = 3.88GYY40 pKa = 10.9GLAAARR46 pKa = 11.84MYY48 pKa = 11.19ADD50 pKa = 3.59PCGADD55 pKa = 3.75LVPTVYY61 pKa = 10.65SGDD64 pKa = 3.18RR65 pKa = 11.84GYY67 pKa = 10.78INRR70 pKa = 11.84FVANASVAITAGDD83 pKa = 3.44TSLILLIKK91 pKa = 10.01PGNQTAFFRR100 pKa = 11.84ADD102 pKa = 3.58TAPTTPGAIGYY113 pKa = 8.69DD114 pKa = 3.2NSIFPGRR121 pKa = 11.84SFYY124 pKa = 10.8TSNASKK130 pKa = 10.47ARR132 pKa = 11.84CAGYY136 pKa = 10.11CINVRR141 pKa = 11.84PNSAPNNATGTIYY154 pKa = 10.6FGNINASAVPNLASITASGLIPLLSEE180 pKa = 4.77SVSCSQALMQPLEE193 pKa = 4.6VKK195 pKa = 9.69WSPGGFDD202 pKa = 4.8DD203 pKa = 5.45RR204 pKa = 11.84YY205 pKa = 9.18NTIGTITDD213 pKa = 3.78DD214 pKa = 4.27DD215 pKa = 4.35SDD217 pKa = 5.02RR218 pKa = 11.84NVLCIVVLGLPAASGMQIRR237 pKa = 11.84QTVITEE243 pKa = 4.18WSPANALTVTNDD255 pKa = 2.79ATAVKK260 pKa = 10.1PSVCDD265 pKa = 3.29KK266 pKa = 11.31DD267 pKa = 4.55CILRR271 pKa = 11.84HH272 pKa = 6.14LKK274 pKa = 10.59RR275 pKa = 11.84KK276 pKa = 10.25DD277 pKa = 3.5SAWWWKK283 pKa = 10.32LGHH286 pKa = 6.57KK287 pKa = 10.23VLDD290 pKa = 3.67VGKK293 pKa = 9.35SVGMGYY299 pKa = 8.19YY300 pKa = 9.81TGGAVGALGAAVRR313 pKa = 11.84FMM315 pKa = 4.85
MM1 pKa = 7.87KK2 pKa = 10.2YY3 pKa = 11.09NLMTKK8 pKa = 10.1SKK10 pKa = 9.14TKK12 pKa = 10.66SNQKK16 pKa = 8.64PAQRR20 pKa = 11.84AKK22 pKa = 10.31PRR24 pKa = 11.84RR25 pKa = 11.84SRR27 pKa = 11.84QRR29 pKa = 11.84ANKK32 pKa = 8.91VQDD35 pKa = 3.52GLDD38 pKa = 3.88GYY40 pKa = 10.9GLAAARR46 pKa = 11.84MYY48 pKa = 11.19ADD50 pKa = 3.59PCGADD55 pKa = 3.75LVPTVYY61 pKa = 10.65SGDD64 pKa = 3.18RR65 pKa = 11.84GYY67 pKa = 10.78INRR70 pKa = 11.84FVANASVAITAGDD83 pKa = 3.44TSLILLIKK91 pKa = 10.01PGNQTAFFRR100 pKa = 11.84ADD102 pKa = 3.58TAPTTPGAIGYY113 pKa = 8.69DD114 pKa = 3.2NSIFPGRR121 pKa = 11.84SFYY124 pKa = 10.8TSNASKK130 pKa = 10.47ARR132 pKa = 11.84CAGYY136 pKa = 10.11CINVRR141 pKa = 11.84PNSAPNNATGTIYY154 pKa = 10.6FGNINASAVPNLASITASGLIPLLSEE180 pKa = 4.77SVSCSQALMQPLEE193 pKa = 4.6VKK195 pKa = 9.69WSPGGFDD202 pKa = 4.8DD203 pKa = 5.45RR204 pKa = 11.84YY205 pKa = 9.18NTIGTITDD213 pKa = 3.78DD214 pKa = 4.27DD215 pKa = 4.35SDD217 pKa = 5.02RR218 pKa = 11.84NVLCIVVLGLPAASGMQIRR237 pKa = 11.84QTVITEE243 pKa = 4.18WSPANALTVTNDD255 pKa = 2.79ATAVKK260 pKa = 10.1PSVCDD265 pKa = 3.29KK266 pKa = 11.31DD267 pKa = 4.55CILRR271 pKa = 11.84HH272 pKa = 6.14LKK274 pKa = 10.59RR275 pKa = 11.84KK276 pKa = 10.25DD277 pKa = 3.5SAWWWKK283 pKa = 10.32LGHH286 pKa = 6.57KK287 pKa = 10.23VLDD290 pKa = 3.67VGKK293 pKa = 9.35SVGMGYY299 pKa = 8.19YY300 pKa = 9.81TGGAVGALGAAVRR313 pKa = 11.84FMM315 pKa = 4.85
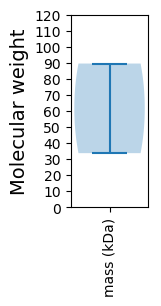
Molecular weight: 33.66 kDa
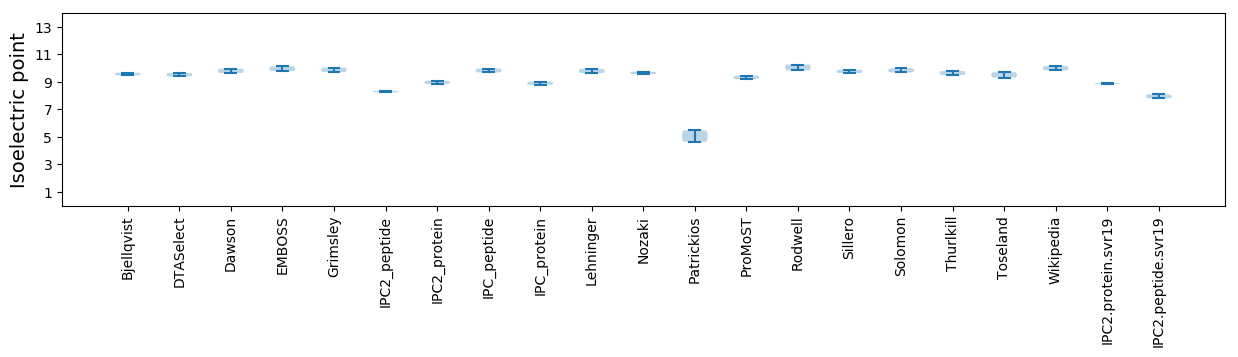
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIL2|A0A1L3KIL2_9VIRU RNA-dependent RNA polymerase OS=Sanxia narna-like virus 1 OX=1923360 PE=4 SV=1
MM1 pKa = 6.93TPQMRR6 pKa = 11.84EE7 pKa = 3.93HH8 pKa = 6.44NSARR12 pKa = 11.84SRR14 pKa = 11.84APRR17 pKa = 11.84MSKK20 pKa = 10.56DD21 pKa = 3.05MHH23 pKa = 6.6LRR25 pKa = 11.84SPLNRR30 pKa = 11.84GPKK33 pKa = 10.07GADD36 pKa = 2.98FLVRR40 pKa = 11.84IPEE43 pKa = 4.22LVADD47 pKa = 4.59PVQVSKK53 pKa = 11.23LSAKK57 pKa = 9.69GVWHH61 pKa = 6.86KK62 pKa = 11.23SDD64 pKa = 3.35NALVEE69 pKa = 4.13TFRR72 pKa = 11.84LIGYY76 pKa = 7.09EE77 pKa = 4.23FKK79 pKa = 11.04NVLCPSPPTSSVPEE93 pKa = 4.03VMSVLKK99 pKa = 10.7NWVAFWLPWFLGDD112 pKa = 4.06DD113 pKa = 3.88TPPQRR118 pKa = 11.84FNPLSLCSPGFRR130 pKa = 11.84SFIRR134 pKa = 11.84NRR136 pKa = 11.84KK137 pKa = 5.37TSKK140 pKa = 8.77TGKK143 pKa = 8.9SRR145 pKa = 11.84KK146 pKa = 9.08IRR148 pKa = 11.84IGALLLYY155 pKa = 9.99SKK157 pKa = 10.74RR158 pKa = 11.84LFPSFTGEE166 pKa = 3.91MVRR169 pKa = 11.84EE170 pKa = 3.99KK171 pKa = 10.98VKK173 pKa = 10.82EE174 pKa = 3.95FGDD177 pKa = 3.49AVTRR181 pKa = 11.84VSPGDD186 pKa = 3.52LPRR189 pKa = 11.84KK190 pKa = 8.54RR191 pKa = 11.84KK192 pKa = 8.78MYY194 pKa = 10.79GSIEE198 pKa = 3.92QSIDD202 pKa = 2.97EE203 pKa = 4.29FLQGGRR209 pKa = 11.84MVADD213 pKa = 3.43YY214 pKa = 10.28SKK216 pKa = 10.59PFAPSTSACYY226 pKa = 9.63EE227 pKa = 3.77KK228 pKa = 10.87SRR230 pKa = 11.84AEE232 pKa = 4.01GGLQRR237 pKa = 11.84FVADD241 pKa = 4.45NILQEE246 pKa = 4.43TVEE249 pKa = 4.36SRR251 pKa = 11.84FFYY254 pKa = 10.82DD255 pKa = 4.68DD256 pKa = 4.91LNHH259 pKa = 6.37IWKK262 pKa = 10.32CSEE265 pKa = 4.31DD266 pKa = 4.05DD267 pKa = 3.79LTFGTPIGLWNEE279 pKa = 3.96FLDD282 pKa = 3.73IMFRR286 pKa = 11.84RR287 pKa = 11.84AMSEE291 pKa = 3.59TGLPDD296 pKa = 3.69QEE298 pKa = 4.43WKK300 pKa = 10.5RR301 pKa = 11.84LPVQATGLTEE311 pKa = 3.93PLKK314 pKa = 11.01VRR316 pKa = 11.84IVTKK320 pKa = 10.42SNWFLQLLTPIQKK333 pKa = 9.54AWHH336 pKa = 5.81GAMRR340 pKa = 11.84SHH342 pKa = 8.26PIYY345 pKa = 10.45QLIGGIPVEE354 pKa = 4.11QALLGLDD361 pKa = 3.79LQRR364 pKa = 11.84KK365 pKa = 7.08QKK367 pKa = 10.27VVSGDD372 pKa = 3.3YY373 pKa = 10.71SAATDD378 pKa = 4.13NIFLEE383 pKa = 4.38YY384 pKa = 10.23TEE386 pKa = 4.53YY387 pKa = 10.87AAKK390 pKa = 10.82AMLEE394 pKa = 3.97RR395 pKa = 11.84TDD397 pKa = 5.67FSFLDD402 pKa = 3.68PQLVQFVPWIKK413 pKa = 10.64KK414 pKa = 9.71LVIHH418 pKa = 5.79SLVRR422 pKa = 11.84SSLEE426 pKa = 3.94LKK428 pKa = 10.72GMEE431 pKa = 4.66PKK433 pKa = 10.65DD434 pKa = 3.27ITRR437 pKa = 11.84GQMMGHH443 pKa = 6.87ILSFPLLCLINRR455 pKa = 11.84SASVMAIPRR464 pKa = 11.84EE465 pKa = 3.56RR466 pKa = 11.84FMRR469 pKa = 11.84INGDD473 pKa = 3.51DD474 pKa = 3.6VLFPANPTEE483 pKa = 3.79YY484 pKa = 10.17RR485 pKa = 11.84KK486 pKa = 9.42WKK488 pKa = 10.05RR489 pKa = 11.84ATASVGLEE497 pKa = 3.59FSLGKK502 pKa = 10.21NYY504 pKa = 10.31YY505 pKa = 10.29SRR507 pKa = 11.84DD508 pKa = 3.39LALVNSTYY516 pKa = 10.73CVWDD520 pKa = 3.68KK521 pKa = 11.68ASARR525 pKa = 11.84WKK527 pKa = 10.13VLSVPNVGLLNMPMEE542 pKa = 4.34KK543 pKa = 10.19QIDD546 pKa = 4.16TEE548 pKa = 4.19TGRR551 pKa = 11.84QIMPWEE557 pKa = 3.96VLAQNWRR564 pKa = 11.84EE565 pKa = 3.76FSRR568 pKa = 11.84FSTKK572 pKa = 8.66KK573 pKa = 7.22TQSTFVRR580 pKa = 11.84LFLKK584 pKa = 10.18HH585 pKa = 5.77YY586 pKa = 8.58PILRR590 pKa = 11.84GFPGPIFGPTQWGGLGAPIPDD611 pKa = 2.98GHH613 pKa = 6.73KK614 pKa = 9.4FTRR617 pKa = 11.84NQLMWMNAHH626 pKa = 6.77RR627 pKa = 11.84LGMFSYY633 pKa = 10.74LQGTRR638 pKa = 11.84TDD640 pKa = 3.55YY641 pKa = 11.45SRR643 pKa = 11.84LSEE646 pKa = 4.35MYY648 pKa = 9.2AQQIEE653 pKa = 4.66KK654 pKa = 9.82YY655 pKa = 8.13VTGIYY660 pKa = 8.88EE661 pKa = 3.94WRR663 pKa = 11.84EE664 pKa = 3.69PEE666 pKa = 4.13YY667 pKa = 11.11GDD669 pKa = 3.93SFGPLEE675 pKa = 4.8SGVFMDD681 pKa = 5.75PYY683 pKa = 11.05SKK685 pKa = 10.92DD686 pKa = 2.7GGYY689 pKa = 10.56AGQVMALRR697 pKa = 11.84RR698 pKa = 11.84WVVDD702 pKa = 3.82ASSMKK707 pKa = 10.05HH708 pKa = 5.02IKK710 pKa = 10.14IFGARR715 pKa = 11.84RR716 pKa = 11.84WNQYY720 pKa = 10.18KK721 pKa = 10.72LSLKK725 pKa = 9.87TGIPPLPQFYY735 pKa = 10.46LDD737 pKa = 4.09KK738 pKa = 10.92LRR740 pKa = 11.84SGDD743 pKa = 3.15IYY745 pKa = 10.81FPRR748 pKa = 11.84PGWNRR753 pKa = 11.84SRR755 pKa = 11.84DD756 pKa = 3.78MTGSRR761 pKa = 11.84YY762 pKa = 8.91EE763 pKa = 3.98DD764 pKa = 3.45RR765 pKa = 11.84ATYY768 pKa = 10.01LHH770 pKa = 7.07EE771 pKa = 4.81IFPSTLKK778 pKa = 10.91NN779 pKa = 3.34
MM1 pKa = 6.93TPQMRR6 pKa = 11.84EE7 pKa = 3.93HH8 pKa = 6.44NSARR12 pKa = 11.84SRR14 pKa = 11.84APRR17 pKa = 11.84MSKK20 pKa = 10.56DD21 pKa = 3.05MHH23 pKa = 6.6LRR25 pKa = 11.84SPLNRR30 pKa = 11.84GPKK33 pKa = 10.07GADD36 pKa = 2.98FLVRR40 pKa = 11.84IPEE43 pKa = 4.22LVADD47 pKa = 4.59PVQVSKK53 pKa = 11.23LSAKK57 pKa = 9.69GVWHH61 pKa = 6.86KK62 pKa = 11.23SDD64 pKa = 3.35NALVEE69 pKa = 4.13TFRR72 pKa = 11.84LIGYY76 pKa = 7.09EE77 pKa = 4.23FKK79 pKa = 11.04NVLCPSPPTSSVPEE93 pKa = 4.03VMSVLKK99 pKa = 10.7NWVAFWLPWFLGDD112 pKa = 4.06DD113 pKa = 3.88TPPQRR118 pKa = 11.84FNPLSLCSPGFRR130 pKa = 11.84SFIRR134 pKa = 11.84NRR136 pKa = 11.84KK137 pKa = 5.37TSKK140 pKa = 8.77TGKK143 pKa = 8.9SRR145 pKa = 11.84KK146 pKa = 9.08IRR148 pKa = 11.84IGALLLYY155 pKa = 9.99SKK157 pKa = 10.74RR158 pKa = 11.84LFPSFTGEE166 pKa = 3.91MVRR169 pKa = 11.84EE170 pKa = 3.99KK171 pKa = 10.98VKK173 pKa = 10.82EE174 pKa = 3.95FGDD177 pKa = 3.49AVTRR181 pKa = 11.84VSPGDD186 pKa = 3.52LPRR189 pKa = 11.84KK190 pKa = 8.54RR191 pKa = 11.84KK192 pKa = 8.78MYY194 pKa = 10.79GSIEE198 pKa = 3.92QSIDD202 pKa = 2.97EE203 pKa = 4.29FLQGGRR209 pKa = 11.84MVADD213 pKa = 3.43YY214 pKa = 10.28SKK216 pKa = 10.59PFAPSTSACYY226 pKa = 9.63EE227 pKa = 3.77KK228 pKa = 10.87SRR230 pKa = 11.84AEE232 pKa = 4.01GGLQRR237 pKa = 11.84FVADD241 pKa = 4.45NILQEE246 pKa = 4.43TVEE249 pKa = 4.36SRR251 pKa = 11.84FFYY254 pKa = 10.82DD255 pKa = 4.68DD256 pKa = 4.91LNHH259 pKa = 6.37IWKK262 pKa = 10.32CSEE265 pKa = 4.31DD266 pKa = 4.05DD267 pKa = 3.79LTFGTPIGLWNEE279 pKa = 3.96FLDD282 pKa = 3.73IMFRR286 pKa = 11.84RR287 pKa = 11.84AMSEE291 pKa = 3.59TGLPDD296 pKa = 3.69QEE298 pKa = 4.43WKK300 pKa = 10.5RR301 pKa = 11.84LPVQATGLTEE311 pKa = 3.93PLKK314 pKa = 11.01VRR316 pKa = 11.84IVTKK320 pKa = 10.42SNWFLQLLTPIQKK333 pKa = 9.54AWHH336 pKa = 5.81GAMRR340 pKa = 11.84SHH342 pKa = 8.26PIYY345 pKa = 10.45QLIGGIPVEE354 pKa = 4.11QALLGLDD361 pKa = 3.79LQRR364 pKa = 11.84KK365 pKa = 7.08QKK367 pKa = 10.27VVSGDD372 pKa = 3.3YY373 pKa = 10.71SAATDD378 pKa = 4.13NIFLEE383 pKa = 4.38YY384 pKa = 10.23TEE386 pKa = 4.53YY387 pKa = 10.87AAKK390 pKa = 10.82AMLEE394 pKa = 3.97RR395 pKa = 11.84TDD397 pKa = 5.67FSFLDD402 pKa = 3.68PQLVQFVPWIKK413 pKa = 10.64KK414 pKa = 9.71LVIHH418 pKa = 5.79SLVRR422 pKa = 11.84SSLEE426 pKa = 3.94LKK428 pKa = 10.72GMEE431 pKa = 4.66PKK433 pKa = 10.65DD434 pKa = 3.27ITRR437 pKa = 11.84GQMMGHH443 pKa = 6.87ILSFPLLCLINRR455 pKa = 11.84SASVMAIPRR464 pKa = 11.84EE465 pKa = 3.56RR466 pKa = 11.84FMRR469 pKa = 11.84INGDD473 pKa = 3.51DD474 pKa = 3.6VLFPANPTEE483 pKa = 3.79YY484 pKa = 10.17RR485 pKa = 11.84KK486 pKa = 9.42WKK488 pKa = 10.05RR489 pKa = 11.84ATASVGLEE497 pKa = 3.59FSLGKK502 pKa = 10.21NYY504 pKa = 10.31YY505 pKa = 10.29SRR507 pKa = 11.84DD508 pKa = 3.39LALVNSTYY516 pKa = 10.73CVWDD520 pKa = 3.68KK521 pKa = 11.68ASARR525 pKa = 11.84WKK527 pKa = 10.13VLSVPNVGLLNMPMEE542 pKa = 4.34KK543 pKa = 10.19QIDD546 pKa = 4.16TEE548 pKa = 4.19TGRR551 pKa = 11.84QIMPWEE557 pKa = 3.96VLAQNWRR564 pKa = 11.84EE565 pKa = 3.76FSRR568 pKa = 11.84FSTKK572 pKa = 8.66KK573 pKa = 7.22TQSTFVRR580 pKa = 11.84LFLKK584 pKa = 10.18HH585 pKa = 5.77YY586 pKa = 8.58PILRR590 pKa = 11.84GFPGPIFGPTQWGGLGAPIPDD611 pKa = 2.98GHH613 pKa = 6.73KK614 pKa = 9.4FTRR617 pKa = 11.84NQLMWMNAHH626 pKa = 6.77RR627 pKa = 11.84LGMFSYY633 pKa = 10.74LQGTRR638 pKa = 11.84TDD640 pKa = 3.55YY641 pKa = 11.45SRR643 pKa = 11.84LSEE646 pKa = 4.35MYY648 pKa = 9.2AQQIEE653 pKa = 4.66KK654 pKa = 9.82YY655 pKa = 8.13VTGIYY660 pKa = 8.88EE661 pKa = 3.94WRR663 pKa = 11.84EE664 pKa = 3.69PEE666 pKa = 4.13YY667 pKa = 11.11GDD669 pKa = 3.93SFGPLEE675 pKa = 4.8SGVFMDD681 pKa = 5.75PYY683 pKa = 11.05SKK685 pKa = 10.92DD686 pKa = 2.7GGYY689 pKa = 10.56AGQVMALRR697 pKa = 11.84RR698 pKa = 11.84WVVDD702 pKa = 3.82ASSMKK707 pKa = 10.05HH708 pKa = 5.02IKK710 pKa = 10.14IFGARR715 pKa = 11.84RR716 pKa = 11.84WNQYY720 pKa = 10.18KK721 pKa = 10.72LSLKK725 pKa = 9.87TGIPPLPQFYY735 pKa = 10.46LDD737 pKa = 4.09KK738 pKa = 10.92LRR740 pKa = 11.84SGDD743 pKa = 3.15IYY745 pKa = 10.81FPRR748 pKa = 11.84PGWNRR753 pKa = 11.84SRR755 pKa = 11.84DD756 pKa = 3.78MTGSRR761 pKa = 11.84YY762 pKa = 8.91EE763 pKa = 3.98DD764 pKa = 3.45RR765 pKa = 11.84ATYY768 pKa = 10.01LHH770 pKa = 7.07EE771 pKa = 4.81IFPSTLKK778 pKa = 10.91NN779 pKa = 3.34
Molecular weight: 89.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1094 |
315 |
779 |
547.0 |
61.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.13 ± 2.309 | 1.188 ± 0.517 |
5.21 ± 0.411 | 3.931 ± 1.49 |
4.57 ± 1.016 | 7.495 ± 0.697 |
1.371 ± 0.368 | 4.845 ± 0.276 |
5.941 ± 0.272 | 8.867 ± 0.942 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.199 ± 0.489 | 4.022 ± 1.005 |
6.307 ± 0.455 | 3.565 ± 0.354 |
7.13 ± 0.708 | 7.861 ± 0.038 |
5.484 ± 0.75 | 5.85 ± 0.409 |
2.377 ± 0.395 | 3.656 ± 0.077 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
