
Candidatus Nasuia deltocephalinicola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Nasuia
Average proteome isoelectric point is 8.8
Get precalculated fractions of proteins
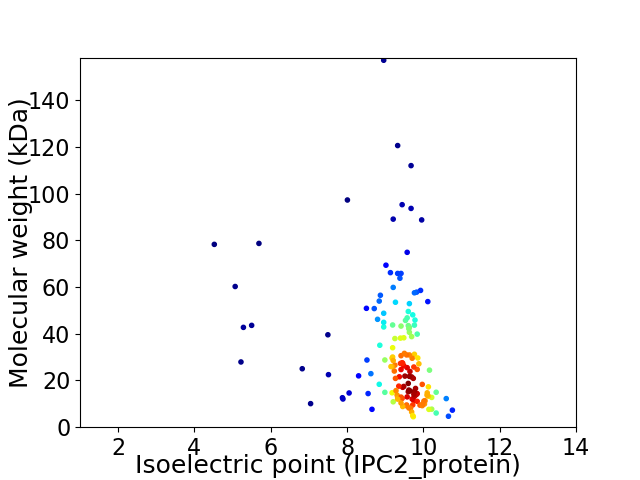
Virtual 2D-PAGE plot for 159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
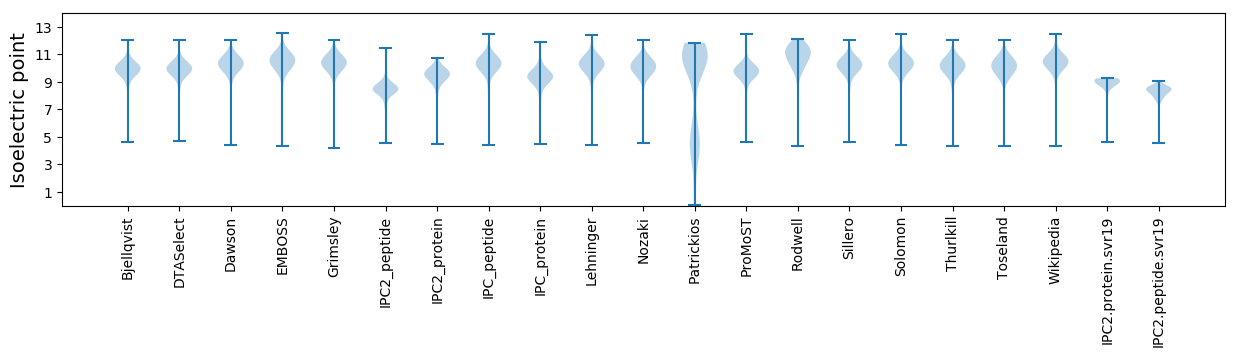
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286QUW8|A0A286QUW8_9PROT Sulfite reductase [NADPH] hemoprotein beta-component OS=Candidatus Nasuia deltocephalinicola OX=1160784 GN=CA212_118 PE=4 SV=1
MM1 pKa = 7.4SKK3 pKa = 10.24IIGIDD8 pKa = 3.1LGTTNSCVAFMDD20 pKa = 4.29GDD22 pKa = 3.81QVKK25 pKa = 10.6VIEE28 pKa = 4.28NSEE31 pKa = 4.1GSRR34 pKa = 11.84ITPSVVAYY42 pKa = 10.14LPNNEE47 pKa = 4.22ILVGIPAKK55 pKa = 10.14RR56 pKa = 11.84QSITNPKK63 pKa = 7.33NTIYY67 pKa = 10.52AVKK70 pKa = 10.17RR71 pKa = 11.84IIGRR75 pKa = 11.84RR76 pKa = 11.84FLEE79 pKa = 4.52KK80 pKa = 10.23EE81 pKa = 3.84VQKK84 pKa = 11.04NINSVPYY91 pKa = 10.35DD92 pKa = 3.19IVEE95 pKa = 4.47ADD97 pKa = 5.48NGDD100 pKa = 3.09AWIKK104 pKa = 10.36IDD106 pKa = 3.9EE107 pKa = 4.55KK108 pKa = 11.04KK109 pKa = 10.22IAPQQISAEE118 pKa = 4.13ILKK121 pKa = 10.55KK122 pKa = 10.19MKK124 pKa = 9.55FTVEE128 pKa = 4.27EE129 pKa = 4.18YY130 pKa = 10.49LGSNIEE136 pKa = 4.17EE137 pKa = 4.1AVVTVPAYY145 pKa = 10.72FNDD148 pKa = 3.76SQRR151 pKa = 11.84QATKK155 pKa = 10.65DD156 pKa = 3.08AGLIAGLNIKK166 pKa = 10.27RR167 pKa = 11.84IINEE171 pKa = 3.61PTAAALAFGLDD182 pKa = 3.31KK183 pKa = 11.47VSINGSKK190 pKa = 9.88IVIYY194 pKa = 10.45DD195 pKa = 3.79LGGGTFDD202 pKa = 3.9ISIIEE207 pKa = 3.84ITEE210 pKa = 3.64IDD212 pKa = 3.74GEE214 pKa = 4.39KK215 pKa = 10.3QFEE218 pKa = 4.19VLATNGDD225 pKa = 3.87TFLGGEE231 pKa = 4.69DD232 pKa = 3.41FDD234 pKa = 4.12QRR236 pKa = 11.84IINYY240 pKa = 8.93VLEE243 pKa = 4.48KK244 pKa = 10.59FKK246 pKa = 11.03EE247 pKa = 4.11DD248 pKa = 3.81NGIDD252 pKa = 3.62LSKK255 pKa = 11.2DD256 pKa = 3.14VLALQRR262 pKa = 11.84LKK264 pKa = 11.02DD265 pKa = 3.65VSEE268 pKa = 4.02KK269 pKa = 11.19AKK271 pKa = 10.57IEE273 pKa = 4.12LSTILQTDD281 pKa = 4.06LSLPYY286 pKa = 9.09ITADD290 pKa = 2.99AYY292 pKa = 10.68GPKK295 pKa = 9.8HH296 pKa = 6.52LNYY299 pKa = 10.17KK300 pKa = 9.86INRR303 pKa = 11.84STLEE307 pKa = 3.85SLVIDD312 pKa = 5.36LIEE315 pKa = 4.43STLKK319 pKa = 10.18PCEE322 pKa = 3.76IAIKK326 pKa = 10.51DD327 pKa = 3.55SGINISDD334 pKa = 3.47IKK336 pKa = 11.13DD337 pKa = 3.51VILVGGQTRR346 pKa = 11.84MPKK349 pKa = 10.22VIQKK353 pKa = 9.95VKK355 pKa = 10.53EE356 pKa = 4.11FFSKK360 pKa = 10.75DD361 pKa = 3.07PRR363 pKa = 11.84RR364 pKa = 11.84DD365 pKa = 3.34INPDD369 pKa = 2.88EE370 pKa = 4.33AVAIGAAIQGQVLSGEE386 pKa = 4.34RR387 pKa = 11.84KK388 pKa = 10.26DD389 pKa = 4.72ILLLDD394 pKa = 3.86VTPLSLGIEE403 pKa = 4.29TLGGVMTKK411 pKa = 9.95MINKK415 pKa = 7.12NTTIPTRR422 pKa = 11.84YY423 pKa = 8.35SQVYY427 pKa = 7.96STADD431 pKa = 3.54DD432 pKa = 3.85NQSSVTIRR440 pKa = 11.84VYY442 pKa = 10.49QGEE445 pKa = 4.42RR446 pKa = 11.84DD447 pKa = 3.55MALHH451 pKa = 6.36NKK453 pKa = 9.29LLAEE457 pKa = 4.93FSLEE461 pKa = 4.38GIPPAARR468 pKa = 11.84GVPQIEE474 pKa = 4.23VTFDD478 pKa = 2.94IDD480 pKa = 4.14SNGILSVKK488 pKa = 10.62AKK490 pKa = 10.8DD491 pKa = 3.6KK492 pKa = 10.6LTLKK496 pKa = 10.03EE497 pKa = 3.88NKK499 pKa = 8.43ITIKK503 pKa = 10.92ANTGLSDD510 pKa = 3.83AEE512 pKa = 3.89IDD514 pKa = 4.81KK515 pKa = 10.15MIKK518 pKa = 10.14DD519 pKa = 3.76AEE521 pKa = 4.41LNMEE525 pKa = 4.15EE526 pKa = 4.21DD527 pKa = 3.72KK528 pKa = 11.05KK529 pKa = 11.02IKK531 pKa = 10.26EE532 pKa = 4.24LTEE535 pKa = 4.12SRR537 pKa = 11.84NKK539 pKa = 10.19AQEE542 pKa = 4.32LIYY545 pKa = 9.63NTKK548 pKa = 10.48KK549 pKa = 10.84SLNEE553 pKa = 4.06YY554 pKa = 10.3GNKK557 pKa = 9.64LQPNEE562 pKa = 4.39LSDD565 pKa = 3.95INLKK569 pKa = 10.6INDD572 pKa = 4.28LEE574 pKa = 4.36ILLSNVKK581 pKa = 8.3STKK584 pKa = 10.26SEE586 pKa = 3.17IDD588 pKa = 3.45YY589 pKa = 11.09KK590 pKa = 11.1VDD592 pKa = 3.38EE593 pKa = 4.85LLKK596 pKa = 10.59SSQKK600 pKa = 8.74MGEE603 pKa = 4.28RR604 pKa = 11.84IYY606 pKa = 11.26SPNSSNNYY614 pKa = 10.03SDD616 pKa = 3.42INNNFDD622 pKa = 3.55INNNDD627 pKa = 3.43FSRR630 pKa = 11.84DD631 pKa = 3.33NNYY634 pKa = 9.43TGNNDD639 pKa = 3.24EE640 pKa = 5.24NSNDD644 pKa = 3.86KK645 pKa = 11.47NNDD648 pKa = 3.19EE649 pKa = 5.21NNDD652 pKa = 3.57EE653 pKa = 5.07NNDD656 pKa = 3.65EE657 pKa = 5.07NNDD660 pKa = 3.65EE661 pKa = 5.07NNDD664 pKa = 3.65EE665 pKa = 5.07NNDD668 pKa = 3.65EE669 pKa = 5.07NNDD672 pKa = 3.65EE673 pKa = 5.07NNDD676 pKa = 3.65EE677 pKa = 5.07NNDD680 pKa = 3.65EE681 pKa = 5.07NNDD684 pKa = 3.73EE685 pKa = 4.43NNDD688 pKa = 4.11IIDD691 pKa = 4.3NYY693 pKa = 10.87DD694 pKa = 2.96NSYY697 pKa = 11.12LEE699 pKa = 4.33KK700 pKa = 10.68KK701 pKa = 10.18
MM1 pKa = 7.4SKK3 pKa = 10.24IIGIDD8 pKa = 3.1LGTTNSCVAFMDD20 pKa = 4.29GDD22 pKa = 3.81QVKK25 pKa = 10.6VIEE28 pKa = 4.28NSEE31 pKa = 4.1GSRR34 pKa = 11.84ITPSVVAYY42 pKa = 10.14LPNNEE47 pKa = 4.22ILVGIPAKK55 pKa = 10.14RR56 pKa = 11.84QSITNPKK63 pKa = 7.33NTIYY67 pKa = 10.52AVKK70 pKa = 10.17RR71 pKa = 11.84IIGRR75 pKa = 11.84RR76 pKa = 11.84FLEE79 pKa = 4.52KK80 pKa = 10.23EE81 pKa = 3.84VQKK84 pKa = 11.04NINSVPYY91 pKa = 10.35DD92 pKa = 3.19IVEE95 pKa = 4.47ADD97 pKa = 5.48NGDD100 pKa = 3.09AWIKK104 pKa = 10.36IDD106 pKa = 3.9EE107 pKa = 4.55KK108 pKa = 11.04KK109 pKa = 10.22IAPQQISAEE118 pKa = 4.13ILKK121 pKa = 10.55KK122 pKa = 10.19MKK124 pKa = 9.55FTVEE128 pKa = 4.27EE129 pKa = 4.18YY130 pKa = 10.49LGSNIEE136 pKa = 4.17EE137 pKa = 4.1AVVTVPAYY145 pKa = 10.72FNDD148 pKa = 3.76SQRR151 pKa = 11.84QATKK155 pKa = 10.65DD156 pKa = 3.08AGLIAGLNIKK166 pKa = 10.27RR167 pKa = 11.84IINEE171 pKa = 3.61PTAAALAFGLDD182 pKa = 3.31KK183 pKa = 11.47VSINGSKK190 pKa = 9.88IVIYY194 pKa = 10.45DD195 pKa = 3.79LGGGTFDD202 pKa = 3.9ISIIEE207 pKa = 3.84ITEE210 pKa = 3.64IDD212 pKa = 3.74GEE214 pKa = 4.39KK215 pKa = 10.3QFEE218 pKa = 4.19VLATNGDD225 pKa = 3.87TFLGGEE231 pKa = 4.69DD232 pKa = 3.41FDD234 pKa = 4.12QRR236 pKa = 11.84IINYY240 pKa = 8.93VLEE243 pKa = 4.48KK244 pKa = 10.59FKK246 pKa = 11.03EE247 pKa = 4.11DD248 pKa = 3.81NGIDD252 pKa = 3.62LSKK255 pKa = 11.2DD256 pKa = 3.14VLALQRR262 pKa = 11.84LKK264 pKa = 11.02DD265 pKa = 3.65VSEE268 pKa = 4.02KK269 pKa = 11.19AKK271 pKa = 10.57IEE273 pKa = 4.12LSTILQTDD281 pKa = 4.06LSLPYY286 pKa = 9.09ITADD290 pKa = 2.99AYY292 pKa = 10.68GPKK295 pKa = 9.8HH296 pKa = 6.52LNYY299 pKa = 10.17KK300 pKa = 9.86INRR303 pKa = 11.84STLEE307 pKa = 3.85SLVIDD312 pKa = 5.36LIEE315 pKa = 4.43STLKK319 pKa = 10.18PCEE322 pKa = 3.76IAIKK326 pKa = 10.51DD327 pKa = 3.55SGINISDD334 pKa = 3.47IKK336 pKa = 11.13DD337 pKa = 3.51VILVGGQTRR346 pKa = 11.84MPKK349 pKa = 10.22VIQKK353 pKa = 9.95VKK355 pKa = 10.53EE356 pKa = 4.11FFSKK360 pKa = 10.75DD361 pKa = 3.07PRR363 pKa = 11.84RR364 pKa = 11.84DD365 pKa = 3.34INPDD369 pKa = 2.88EE370 pKa = 4.33AVAIGAAIQGQVLSGEE386 pKa = 4.34RR387 pKa = 11.84KK388 pKa = 10.26DD389 pKa = 4.72ILLLDD394 pKa = 3.86VTPLSLGIEE403 pKa = 4.29TLGGVMTKK411 pKa = 9.95MINKK415 pKa = 7.12NTTIPTRR422 pKa = 11.84YY423 pKa = 8.35SQVYY427 pKa = 7.96STADD431 pKa = 3.54DD432 pKa = 3.85NQSSVTIRR440 pKa = 11.84VYY442 pKa = 10.49QGEE445 pKa = 4.42RR446 pKa = 11.84DD447 pKa = 3.55MALHH451 pKa = 6.36NKK453 pKa = 9.29LLAEE457 pKa = 4.93FSLEE461 pKa = 4.38GIPPAARR468 pKa = 11.84GVPQIEE474 pKa = 4.23VTFDD478 pKa = 2.94IDD480 pKa = 4.14SNGILSVKK488 pKa = 10.62AKK490 pKa = 10.8DD491 pKa = 3.6KK492 pKa = 10.6LTLKK496 pKa = 10.03EE497 pKa = 3.88NKK499 pKa = 8.43ITIKK503 pKa = 10.92ANTGLSDD510 pKa = 3.83AEE512 pKa = 3.89IDD514 pKa = 4.81KK515 pKa = 10.15MIKK518 pKa = 10.14DD519 pKa = 3.76AEE521 pKa = 4.41LNMEE525 pKa = 4.15EE526 pKa = 4.21DD527 pKa = 3.72KK528 pKa = 11.05KK529 pKa = 11.02IKK531 pKa = 10.26EE532 pKa = 4.24LTEE535 pKa = 4.12SRR537 pKa = 11.84NKK539 pKa = 10.19AQEE542 pKa = 4.32LIYY545 pKa = 9.63NTKK548 pKa = 10.48KK549 pKa = 10.84SLNEE553 pKa = 4.06YY554 pKa = 10.3GNKK557 pKa = 9.64LQPNEE562 pKa = 4.39LSDD565 pKa = 3.95INLKK569 pKa = 10.6INDD572 pKa = 4.28LEE574 pKa = 4.36ILLSNVKK581 pKa = 8.3STKK584 pKa = 10.26SEE586 pKa = 3.17IDD588 pKa = 3.45YY589 pKa = 11.09KK590 pKa = 11.1VDD592 pKa = 3.38EE593 pKa = 4.85LLKK596 pKa = 10.59SSQKK600 pKa = 8.74MGEE603 pKa = 4.28RR604 pKa = 11.84IYY606 pKa = 11.26SPNSSNNYY614 pKa = 10.03SDD616 pKa = 3.42INNNFDD622 pKa = 3.55INNNDD627 pKa = 3.43FSRR630 pKa = 11.84DD631 pKa = 3.33NNYY634 pKa = 9.43TGNNDD639 pKa = 3.24EE640 pKa = 5.24NSNDD644 pKa = 3.86KK645 pKa = 11.47NNDD648 pKa = 3.19EE649 pKa = 5.21NNDD652 pKa = 3.57EE653 pKa = 5.07NNDD656 pKa = 3.65EE657 pKa = 5.07NNDD660 pKa = 3.65EE661 pKa = 5.07NNDD664 pKa = 3.65EE665 pKa = 5.07NNDD668 pKa = 3.65EE669 pKa = 5.07NNDD672 pKa = 3.65EE673 pKa = 5.07NNDD676 pKa = 3.65EE677 pKa = 5.07NNDD680 pKa = 3.65EE681 pKa = 5.07NNDD684 pKa = 3.73EE685 pKa = 4.43NNDD688 pKa = 4.11IIDD691 pKa = 4.3NYY693 pKa = 10.87DD694 pKa = 2.96NSYY697 pKa = 11.12LEE699 pKa = 4.33KK700 pKa = 10.68KK701 pKa = 10.18
Molecular weight: 78.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A248QFN0|A0A248QFN0_9PROT 50S ribosomal protein L27p OS=Candidatus Nasuia deltocephalinicola OX=1160784 GN=CA212_071 PE=3 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84HH3 pKa = 5.45LKK5 pKa = 10.41KK6 pKa = 9.88RR7 pKa = 11.84DD8 pKa = 3.34KK9 pKa = 11.3LGIKK13 pKa = 9.83YY14 pKa = 8.41SHH16 pKa = 6.76KK17 pKa = 10.51KK18 pKa = 10.36SLISNLSNSLIKK30 pKa = 10.68YY31 pKa = 8.01EE32 pKa = 3.91YY33 pKa = 10.93LKK35 pKa = 9.6ITFVRR40 pKa = 11.84AKK42 pKa = 9.6VLKK45 pKa = 10.63SFLEE49 pKa = 4.5PIITVSKK56 pKa = 10.94YY57 pKa = 8.99FFNYY61 pKa = 9.15NVRR64 pKa = 11.84FVFKK68 pKa = 10.55KK69 pKa = 10.29LRR71 pKa = 11.84NTEE74 pKa = 4.15SVLKK78 pKa = 9.73LFKK81 pKa = 10.54IIGPRR86 pKa = 11.84YY87 pKa = 7.77FFRR90 pKa = 11.84NGGYY94 pKa = 9.81LQIFKK99 pKa = 10.67KK100 pKa = 10.56GFRR103 pKa = 11.84RR104 pKa = 11.84GDD106 pKa = 3.63NACLALIKK114 pKa = 10.64LII116 pKa = 4.38
MM1 pKa = 7.58RR2 pKa = 11.84HH3 pKa = 5.45LKK5 pKa = 10.41KK6 pKa = 9.88RR7 pKa = 11.84DD8 pKa = 3.34KK9 pKa = 11.3LGIKK13 pKa = 9.83YY14 pKa = 8.41SHH16 pKa = 6.76KK17 pKa = 10.51KK18 pKa = 10.36SLISNLSNSLIKK30 pKa = 10.68YY31 pKa = 8.01EE32 pKa = 3.91YY33 pKa = 10.93LKK35 pKa = 9.6ITFVRR40 pKa = 11.84AKK42 pKa = 9.6VLKK45 pKa = 10.63SFLEE49 pKa = 4.5PIITVSKK56 pKa = 10.94YY57 pKa = 8.99FFNYY61 pKa = 9.15NVRR64 pKa = 11.84FVFKK68 pKa = 10.55KK69 pKa = 10.29LRR71 pKa = 11.84NTEE74 pKa = 4.15SVLKK78 pKa = 9.73LFKK81 pKa = 10.54IIGPRR86 pKa = 11.84YY87 pKa = 7.77FFRR90 pKa = 11.84NGGYY94 pKa = 9.81LQIFKK99 pKa = 10.67KK100 pKa = 10.56GFRR103 pKa = 11.84RR104 pKa = 11.84GDD106 pKa = 3.63NACLALIKK114 pKa = 10.64LII116 pKa = 4.38
Molecular weight: 13.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
43276 |
37 |
1361 |
272.2 |
32.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.257 ± 0.132 | 1.174 ± 0.056 |
3.358 ± 0.215 | 3.187 ± 0.213 |
9.315 ± 0.503 | 3.632 ± 0.211 |
0.915 ± 0.063 | 15.452 ± 0.266 |
14.424 ± 0.452 | 10.59 ± 0.191 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.135 ± 0.066 | 12.631 ± 0.295 |
1.752 ± 0.085 | 1.109 ± 0.067 |
2.068 ± 0.103 | 6.304 ± 0.216 |
2.308 ± 0.12 | 2.537 ± 0.137 |
0.525 ± 0.061 | 6.329 ± 0.24 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
