
Microbacterium mitrae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
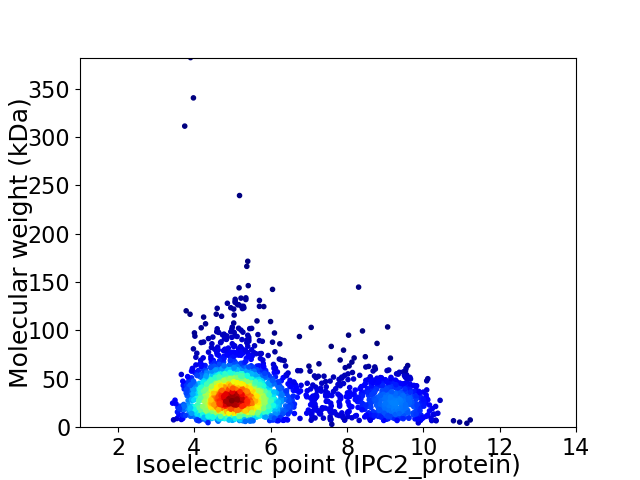
Virtual 2D-PAGE plot for 2481 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C8HJV4|A0A5C8HJV4_9MICO Phosphoribosylformylglycinamidine synthase subunit PurL OS=Microbacterium mitrae OX=664640 GN=purL PE=3 SV=1
MM1 pKa = 8.55SMEE4 pKa = 4.07GRR6 pKa = 11.84FGLKK10 pKa = 9.95AGAVVTFAALIACTSAATTSFAAEE34 pKa = 4.1PTEE37 pKa = 4.1VSIPTPQNLEE47 pKa = 3.9DD48 pKa = 3.28GRR50 pKa = 11.84YY51 pKa = 9.07IVLLDD56 pKa = 4.17DD57 pKa = 5.15APAATYY63 pKa = 10.29EE64 pKa = 4.21GGEE67 pKa = 4.32AGLTATKK74 pKa = 9.7PAEE77 pKa = 4.22GDD79 pKa = 3.56KK80 pKa = 11.28LDD82 pKa = 3.77ATTDD86 pKa = 3.37AVVEE90 pKa = 4.22YY91 pKa = 10.57SSHH94 pKa = 7.24LEE96 pKa = 3.87NLQDD100 pKa = 3.41EE101 pKa = 4.87VAAEE105 pKa = 4.24VGATPDD111 pKa = 3.37ANLTITLNAFTANLTASQAKK131 pKa = 9.13QLAATNGVSAVVPDD145 pKa = 5.13EE146 pKa = 4.24ILHH149 pKa = 5.37PTAVPSTEE157 pKa = 3.88FLGLEE162 pKa = 4.16GDD164 pKa = 3.55NGVWEE169 pKa = 4.86TYY171 pKa = 10.84AGGPEE176 pKa = 4.1GAGEE180 pKa = 4.3GVVVGVIDD188 pKa = 3.79TGIAPEE194 pKa = 4.21NPSFAGEE201 pKa = 4.03PLGTTSGATPYY212 pKa = 11.03LEE214 pKa = 4.37GNTVVFNKK222 pKa = 10.38ADD224 pKa = 3.64GGQFRR229 pKa = 11.84SDD231 pKa = 3.7RR232 pKa = 11.84VTGDD236 pKa = 2.78QWNLNDD242 pKa = 4.25YY243 pKa = 7.43STKK246 pKa = 10.69LIAAHH251 pKa = 5.88YY252 pKa = 8.67FSAGAAASGFDD263 pKa = 3.98FQWDD267 pKa = 3.36ILSPRR272 pKa = 11.84DD273 pKa = 3.59GAGHH277 pKa = 6.87GSHH280 pKa = 6.47TASTAAGNYY289 pKa = 8.58NVEE292 pKa = 4.45TTVSGLDD299 pKa = 3.38FGPISGVAPAAKK311 pKa = 8.79IASYY315 pKa = 8.09KK316 pKa = 8.83TCYY319 pKa = 9.61EE320 pKa = 4.08GQDD323 pKa = 3.67PAVTTDD329 pKa = 4.76DD330 pKa = 3.32ICAGSDD336 pKa = 3.21LLAAIDD342 pKa = 3.94RR343 pKa = 11.84ATKK346 pKa = 10.64DD347 pKa = 3.37GVDD350 pKa = 3.95VINFSIGGGAATTVWDD366 pKa = 4.57ALDD369 pKa = 3.28QAFFNAATAGIFVSASAGNSGPDD392 pKa = 3.71YY393 pKa = 10.21VTADD397 pKa = 4.08HH398 pKa = 7.06ASPWYY403 pKa = 6.52TTVAASTIPTYY414 pKa = 10.58EE415 pKa = 3.81GTVVGEE421 pKa = 4.19GFEE424 pKa = 4.58APGATVSVPQGGEE437 pKa = 3.85VTGDD441 pKa = 3.78AIYY444 pKa = 10.76AGDD447 pKa = 3.74VAASGATAANAALCLPNTLDD467 pKa = 3.43ASLVAGKK474 pKa = 10.05IVVCDD479 pKa = 3.62RR480 pKa = 11.84GSNARR485 pKa = 11.84VDD487 pKa = 3.43KK488 pKa = 10.75SANVQAAGGIGMVLVNVGPGSLDD511 pKa = 3.1TDD513 pKa = 3.58MHH515 pKa = 6.48SVPTVHH521 pKa = 7.49IADD524 pKa = 4.46TYY526 pKa = 10.98RR527 pKa = 11.84ADD529 pKa = 3.88LLATITEE536 pKa = 4.43TPEE539 pKa = 3.53ITLLPQNITDD549 pKa = 4.62FVTPVPQIAGFSSRR563 pKa = 11.84GPMMADD569 pKa = 2.98GSDD572 pKa = 3.03ILKK575 pKa = 10.09PDD577 pKa = 3.14IAAPGVNILAAVNNVEE593 pKa = 4.9DD594 pKa = 5.46AAPQWGLMSGTSMAAPHH611 pKa = 6.44IAGLAALYY619 pKa = 10.43LGEE622 pKa = 4.98RR623 pKa = 11.84PQATPGEE630 pKa = 4.34IKK632 pKa = 10.39SALMTTAYY640 pKa = 8.77DD641 pKa = 3.83TVDD644 pKa = 4.35SEE646 pKa = 5.1DD647 pKa = 4.11ASNPDD652 pKa = 3.73PFAQGAGQVDD662 pKa = 3.39AKK664 pKa = 11.04LYY666 pKa = 10.66FNAGLLYY673 pKa = 10.98LNGPADD679 pKa = 3.19WAAYY683 pKa = 10.52LEE685 pKa = 4.53GKK687 pKa = 9.77GLYY690 pKa = 9.79EE691 pKa = 4.07FDD693 pKa = 3.39GVEE696 pKa = 4.71PIDD699 pKa = 5.08GSDD702 pKa = 3.92LNLASISIGSLAGPQTVTRR721 pKa = 11.84TVTAQTAGTYY731 pKa = 7.66EE732 pKa = 3.88ATIDD736 pKa = 3.76VPGVTTVVEE745 pKa = 4.35PSSITLAAGEE755 pKa = 4.5SATFTVSFEE764 pKa = 4.02NATAPTEE771 pKa = 3.87QWATGFLTWSSDD783 pKa = 3.8TIDD786 pKa = 3.57VRR788 pKa = 11.84SPIAVFPTAADD799 pKa = 3.71APLEE803 pKa = 4.07LYY805 pKa = 11.0GEE807 pKa = 4.49GVEE810 pKa = 5.01GVIDD814 pKa = 3.75YY815 pKa = 10.73SITPGYY821 pKa = 10.61DD822 pKa = 2.98GEE824 pKa = 4.91LPLNLSGLTPYY835 pKa = 10.39VLHH838 pKa = 7.59EE839 pKa = 4.7DD840 pKa = 3.46TDD842 pKa = 4.23NPVPGHH848 pKa = 6.41SGNQDD853 pKa = 3.0SGDD856 pKa = 3.56ADD858 pKa = 3.9GEE860 pKa = 4.1IGFILEE866 pKa = 4.21VQEE869 pKa = 4.23GAEE872 pKa = 4.18LARR875 pKa = 11.84IDD877 pKa = 5.37LDD879 pKa = 3.79SSDD882 pKa = 5.15DD883 pKa = 3.82AGSDD887 pKa = 3.45LDD889 pKa = 3.71LTVYY893 pKa = 10.65HH894 pKa = 6.62LVSPTSYY901 pKa = 10.65DD902 pKa = 3.09KK903 pKa = 11.16KK904 pKa = 11.06YY905 pKa = 10.8DD906 pKa = 3.77AQTGSADD913 pKa = 3.67EE914 pKa = 4.19QVTIHH919 pKa = 6.73APAAGTYY926 pKa = 9.77FILVDD931 pKa = 4.12MYY933 pKa = 11.21SVSGPMTWDD942 pKa = 2.89LTTAVVDD949 pKa = 4.73GAADD953 pKa = 3.85GALTATPNPVAATKK967 pKa = 10.44GVSTPVSLSWAGLEE981 pKa = 3.78NDD983 pKa = 3.36QRR985 pKa = 11.84YY986 pKa = 9.37LGYY989 pKa = 10.25VQYY992 pKa = 11.15GDD994 pKa = 4.01SAVSTIVTVDD1004 pKa = 3.29AGAAAPANTAAPVLSGSGVAGEE1026 pKa = 4.53TLTVTPGEE1034 pKa = 4.03WDD1036 pKa = 3.2QDD1038 pKa = 3.47GLTFKK1043 pKa = 11.33YY1044 pKa = 9.73EE1045 pKa = 3.68WLKK1048 pKa = 11.03NGEE1051 pKa = 4.5AIAVADD1057 pKa = 4.26APAGVNAVVPPDD1069 pKa = 3.54ATDD1072 pKa = 3.02SSTYY1076 pKa = 9.99TPTAADD1082 pKa = 3.29VGATISVRR1090 pKa = 11.84VYY1092 pKa = 10.82ASAEE1096 pKa = 4.04GNPNAGTADD1105 pKa = 3.51SNGIVITAAPTTSPTPTVDD1124 pKa = 3.13PTDD1127 pKa = 3.71PTNPTVAPTTAPTTPGAGGLPVTGAPDD1154 pKa = 3.24ATLPLLVGFLIVALGGAAFVIGRR1177 pKa = 11.84KK1178 pKa = 9.11RR1179 pKa = 11.84RR1180 pKa = 11.84AHH1182 pKa = 6.68AEE1184 pKa = 3.76
MM1 pKa = 8.55SMEE4 pKa = 4.07GRR6 pKa = 11.84FGLKK10 pKa = 9.95AGAVVTFAALIACTSAATTSFAAEE34 pKa = 4.1PTEE37 pKa = 4.1VSIPTPQNLEE47 pKa = 3.9DD48 pKa = 3.28GRR50 pKa = 11.84YY51 pKa = 9.07IVLLDD56 pKa = 4.17DD57 pKa = 5.15APAATYY63 pKa = 10.29EE64 pKa = 4.21GGEE67 pKa = 4.32AGLTATKK74 pKa = 9.7PAEE77 pKa = 4.22GDD79 pKa = 3.56KK80 pKa = 11.28LDD82 pKa = 3.77ATTDD86 pKa = 3.37AVVEE90 pKa = 4.22YY91 pKa = 10.57SSHH94 pKa = 7.24LEE96 pKa = 3.87NLQDD100 pKa = 3.41EE101 pKa = 4.87VAAEE105 pKa = 4.24VGATPDD111 pKa = 3.37ANLTITLNAFTANLTASQAKK131 pKa = 9.13QLAATNGVSAVVPDD145 pKa = 5.13EE146 pKa = 4.24ILHH149 pKa = 5.37PTAVPSTEE157 pKa = 3.88FLGLEE162 pKa = 4.16GDD164 pKa = 3.55NGVWEE169 pKa = 4.86TYY171 pKa = 10.84AGGPEE176 pKa = 4.1GAGEE180 pKa = 4.3GVVVGVIDD188 pKa = 3.79TGIAPEE194 pKa = 4.21NPSFAGEE201 pKa = 4.03PLGTTSGATPYY212 pKa = 11.03LEE214 pKa = 4.37GNTVVFNKK222 pKa = 10.38ADD224 pKa = 3.64GGQFRR229 pKa = 11.84SDD231 pKa = 3.7RR232 pKa = 11.84VTGDD236 pKa = 2.78QWNLNDD242 pKa = 4.25YY243 pKa = 7.43STKK246 pKa = 10.69LIAAHH251 pKa = 5.88YY252 pKa = 8.67FSAGAAASGFDD263 pKa = 3.98FQWDD267 pKa = 3.36ILSPRR272 pKa = 11.84DD273 pKa = 3.59GAGHH277 pKa = 6.87GSHH280 pKa = 6.47TASTAAGNYY289 pKa = 8.58NVEE292 pKa = 4.45TTVSGLDD299 pKa = 3.38FGPISGVAPAAKK311 pKa = 8.79IASYY315 pKa = 8.09KK316 pKa = 8.83TCYY319 pKa = 9.61EE320 pKa = 4.08GQDD323 pKa = 3.67PAVTTDD329 pKa = 4.76DD330 pKa = 3.32ICAGSDD336 pKa = 3.21LLAAIDD342 pKa = 3.94RR343 pKa = 11.84ATKK346 pKa = 10.64DD347 pKa = 3.37GVDD350 pKa = 3.95VINFSIGGGAATTVWDD366 pKa = 4.57ALDD369 pKa = 3.28QAFFNAATAGIFVSASAGNSGPDD392 pKa = 3.71YY393 pKa = 10.21VTADD397 pKa = 4.08HH398 pKa = 7.06ASPWYY403 pKa = 6.52TTVAASTIPTYY414 pKa = 10.58EE415 pKa = 3.81GTVVGEE421 pKa = 4.19GFEE424 pKa = 4.58APGATVSVPQGGEE437 pKa = 3.85VTGDD441 pKa = 3.78AIYY444 pKa = 10.76AGDD447 pKa = 3.74VAASGATAANAALCLPNTLDD467 pKa = 3.43ASLVAGKK474 pKa = 10.05IVVCDD479 pKa = 3.62RR480 pKa = 11.84GSNARR485 pKa = 11.84VDD487 pKa = 3.43KK488 pKa = 10.75SANVQAAGGIGMVLVNVGPGSLDD511 pKa = 3.1TDD513 pKa = 3.58MHH515 pKa = 6.48SVPTVHH521 pKa = 7.49IADD524 pKa = 4.46TYY526 pKa = 10.98RR527 pKa = 11.84ADD529 pKa = 3.88LLATITEE536 pKa = 4.43TPEE539 pKa = 3.53ITLLPQNITDD549 pKa = 4.62FVTPVPQIAGFSSRR563 pKa = 11.84GPMMADD569 pKa = 2.98GSDD572 pKa = 3.03ILKK575 pKa = 10.09PDD577 pKa = 3.14IAAPGVNILAAVNNVEE593 pKa = 4.9DD594 pKa = 5.46AAPQWGLMSGTSMAAPHH611 pKa = 6.44IAGLAALYY619 pKa = 10.43LGEE622 pKa = 4.98RR623 pKa = 11.84PQATPGEE630 pKa = 4.34IKK632 pKa = 10.39SALMTTAYY640 pKa = 8.77DD641 pKa = 3.83TVDD644 pKa = 4.35SEE646 pKa = 5.1DD647 pKa = 4.11ASNPDD652 pKa = 3.73PFAQGAGQVDD662 pKa = 3.39AKK664 pKa = 11.04LYY666 pKa = 10.66FNAGLLYY673 pKa = 10.98LNGPADD679 pKa = 3.19WAAYY683 pKa = 10.52LEE685 pKa = 4.53GKK687 pKa = 9.77GLYY690 pKa = 9.79EE691 pKa = 4.07FDD693 pKa = 3.39GVEE696 pKa = 4.71PIDD699 pKa = 5.08GSDD702 pKa = 3.92LNLASISIGSLAGPQTVTRR721 pKa = 11.84TVTAQTAGTYY731 pKa = 7.66EE732 pKa = 3.88ATIDD736 pKa = 3.76VPGVTTVVEE745 pKa = 4.35PSSITLAAGEE755 pKa = 4.5SATFTVSFEE764 pKa = 4.02NATAPTEE771 pKa = 3.87QWATGFLTWSSDD783 pKa = 3.8TIDD786 pKa = 3.57VRR788 pKa = 11.84SPIAVFPTAADD799 pKa = 3.71APLEE803 pKa = 4.07LYY805 pKa = 11.0GEE807 pKa = 4.49GVEE810 pKa = 5.01GVIDD814 pKa = 3.75YY815 pKa = 10.73SITPGYY821 pKa = 10.61DD822 pKa = 2.98GEE824 pKa = 4.91LPLNLSGLTPYY835 pKa = 10.39VLHH838 pKa = 7.59EE839 pKa = 4.7DD840 pKa = 3.46TDD842 pKa = 4.23NPVPGHH848 pKa = 6.41SGNQDD853 pKa = 3.0SGDD856 pKa = 3.56ADD858 pKa = 3.9GEE860 pKa = 4.1IGFILEE866 pKa = 4.21VQEE869 pKa = 4.23GAEE872 pKa = 4.18LARR875 pKa = 11.84IDD877 pKa = 5.37LDD879 pKa = 3.79SSDD882 pKa = 5.15DD883 pKa = 3.82AGSDD887 pKa = 3.45LDD889 pKa = 3.71LTVYY893 pKa = 10.65HH894 pKa = 6.62LVSPTSYY901 pKa = 10.65DD902 pKa = 3.09KK903 pKa = 11.16KK904 pKa = 11.06YY905 pKa = 10.8DD906 pKa = 3.77AQTGSADD913 pKa = 3.67EE914 pKa = 4.19QVTIHH919 pKa = 6.73APAAGTYY926 pKa = 9.77FILVDD931 pKa = 4.12MYY933 pKa = 11.21SVSGPMTWDD942 pKa = 2.89LTTAVVDD949 pKa = 4.73GAADD953 pKa = 3.85GALTATPNPVAATKK967 pKa = 10.44GVSTPVSLSWAGLEE981 pKa = 3.78NDD983 pKa = 3.36QRR985 pKa = 11.84YY986 pKa = 9.37LGYY989 pKa = 10.25VQYY992 pKa = 11.15GDD994 pKa = 4.01SAVSTIVTVDD1004 pKa = 3.29AGAAAPANTAAPVLSGSGVAGEE1026 pKa = 4.53TLTVTPGEE1034 pKa = 4.03WDD1036 pKa = 3.2QDD1038 pKa = 3.47GLTFKK1043 pKa = 11.33YY1044 pKa = 9.73EE1045 pKa = 3.68WLKK1048 pKa = 11.03NGEE1051 pKa = 4.5AIAVADD1057 pKa = 4.26APAGVNAVVPPDD1069 pKa = 3.54ATDD1072 pKa = 3.02SSTYY1076 pKa = 9.99TPTAADD1082 pKa = 3.29VGATISVRR1090 pKa = 11.84VYY1092 pKa = 10.82ASAEE1096 pKa = 4.04GNPNAGTADD1105 pKa = 3.51SNGIVITAAPTTSPTPTVDD1124 pKa = 3.13PTDD1127 pKa = 3.71PTNPTVAPTTAPTTPGAGGLPVTGAPDD1154 pKa = 3.24ATLPLLVGFLIVALGGAAFVIGRR1177 pKa = 11.84KK1178 pKa = 9.11RR1179 pKa = 11.84RR1180 pKa = 11.84AHH1182 pKa = 6.68AEE1184 pKa = 3.76
Molecular weight: 120.25 kDa
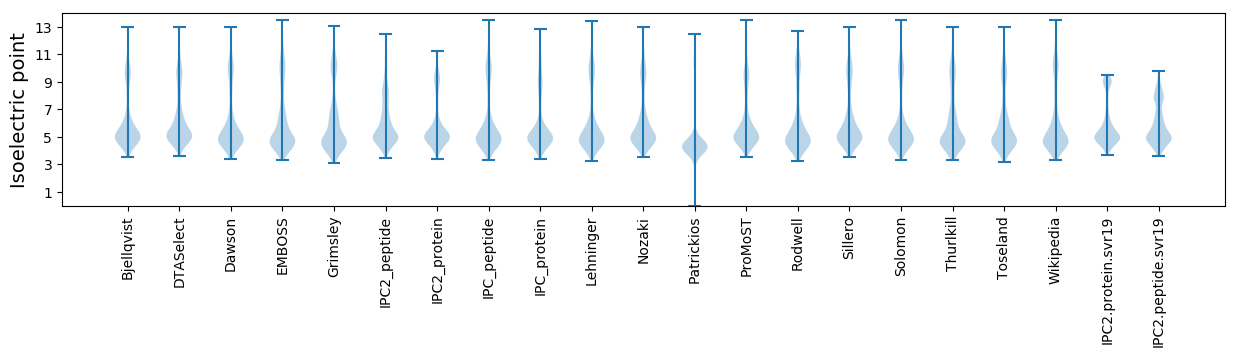
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C8HLN7|A0A5C8HLN7_9MICO Penicillin-insensitive transglycosylase OS=Microbacterium mitrae OX=664640 GN=FVP60_12320 PE=4 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84HH3 pKa = 5.85APSTGTRR10 pKa = 11.84RR11 pKa = 11.84PPSRR15 pKa = 11.84PARR18 pKa = 11.84FRR20 pKa = 11.84PCGLPATPQRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84ARR35 pKa = 11.84VVPSARR41 pKa = 11.84RR42 pKa = 11.84ARR44 pKa = 11.84AVPQGAGRR52 pKa = 11.84RR53 pKa = 11.84PRR55 pKa = 11.84RR56 pKa = 11.84ARR58 pKa = 11.84RR59 pKa = 11.84ARR61 pKa = 11.84WWRR64 pKa = 3.35
MM1 pKa = 7.46RR2 pKa = 11.84HH3 pKa = 5.85APSTGTRR10 pKa = 11.84RR11 pKa = 11.84PPSRR15 pKa = 11.84PARR18 pKa = 11.84FRR20 pKa = 11.84PCGLPATPQRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84ARR35 pKa = 11.84VVPSARR41 pKa = 11.84RR42 pKa = 11.84ARR44 pKa = 11.84AVPQGAGRR52 pKa = 11.84RR53 pKa = 11.84PRR55 pKa = 11.84RR56 pKa = 11.84ARR58 pKa = 11.84RR59 pKa = 11.84ARR61 pKa = 11.84WWRR64 pKa = 3.35
Molecular weight: 7.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
825282 |
29 |
3744 |
332.6 |
35.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.318 ± 0.072 | 0.496 ± 0.015 |
6.055 ± 0.041 | 5.794 ± 0.051 |
3.231 ± 0.03 | 8.417 ± 0.044 |
1.976 ± 0.025 | 5.139 ± 0.038 |
2.336 ± 0.036 | 9.643 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.07 ± 0.024 | 2.361 ± 0.027 |
5.077 ± 0.034 | 3.009 ± 0.025 |
6.662 ± 0.058 | 5.712 ± 0.033 |
6.457 ± 0.066 | 8.745 ± 0.048 |
1.452 ± 0.021 | 2.051 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
