
Microbacterium sp. XT11
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
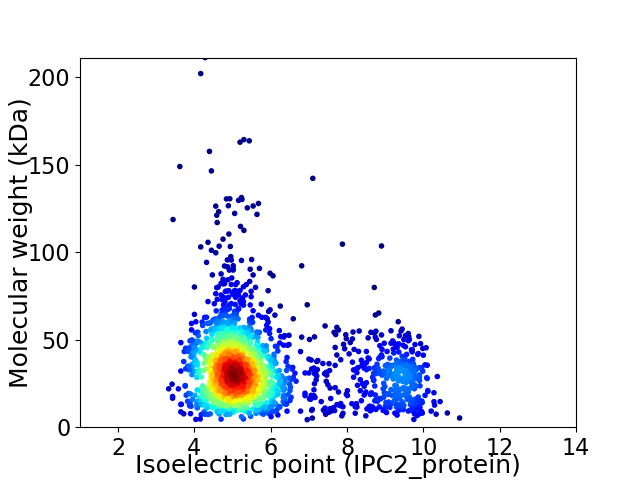
Virtual 2D-PAGE plot for 1927 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4H705|A0A0U4H705_9MICO o-succinylbenzoate synthase OS=Microbacterium sp. XT11 OX=367477 GN=menC PE=3 SV=1
MM1 pKa = 7.06ATSTRR6 pKa = 11.84ALIGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84PAARR17 pKa = 11.84PGRR20 pKa = 11.84SGEE23 pKa = 3.78RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84AGVAAAIVAALAVSLLGVALPSPALAAGNTPPSNAYY62 pKa = 10.06SNQWLYY68 pKa = 11.42AYY70 pKa = 10.33VEE72 pKa = 4.18AGEE75 pKa = 4.53TVVGTGNIEE84 pKa = 4.0QVVAPDD90 pKa = 3.54GTTALGPGTYY100 pKa = 10.34GPAAADD106 pKa = 3.91GVWRR110 pKa = 11.84IHH112 pKa = 6.68LPSQGTSIGYY122 pKa = 8.39SWTVDD127 pKa = 2.64VHH129 pKa = 6.45AAGSGQVIPGRR140 pKa = 11.84VWSDD144 pKa = 2.77RR145 pKa = 11.84YY146 pKa = 11.11YY147 pKa = 9.23MQQASGTANTVDD159 pKa = 3.63LNFWIVNNTGYY170 pKa = 10.98VYY172 pKa = 10.69AVEE175 pKa = 4.49LDD177 pKa = 4.12DD178 pKa = 4.22YY179 pKa = 11.55NGVNSIFEE187 pKa = 4.48ANSVGWADD195 pKa = 4.02EE196 pKa = 4.26ADD198 pKa = 4.9CIPPYY203 pKa = 10.76ASYY206 pKa = 10.4EE207 pKa = 4.05PPGGVGTTLPLPDD220 pKa = 4.66CGEE223 pKa = 4.11TYY225 pKa = 10.43RR226 pKa = 11.84VFFEE230 pKa = 4.84EE231 pKa = 4.7PADD234 pKa = 4.33DD235 pKa = 4.56LPQVSSTNLGSVNVRR250 pKa = 11.84PDD252 pKa = 3.52LLGPADD258 pKa = 4.32LAVDD262 pKa = 4.23DD263 pKa = 5.33LAFAPAGGGSATGTFTYY280 pKa = 10.36SINDD284 pKa = 3.82RR285 pKa = 11.84FTGGYY290 pKa = 9.12VLQIDD295 pKa = 3.75VDD297 pKa = 4.06GDD299 pKa = 3.64GSYY302 pKa = 11.07DD303 pKa = 3.59GQRR306 pKa = 11.84DD307 pKa = 3.18RR308 pKa = 11.84SIRR311 pKa = 11.84LGADD315 pKa = 3.02GSGSYY320 pKa = 10.44SYY322 pKa = 11.59AFDD325 pKa = 4.11GLDD328 pKa = 3.41GLGDD332 pKa = 4.56PIEE335 pKa = 4.84DD336 pKa = 4.05CTRR339 pKa = 11.84MGARR343 pKa = 11.84VFFDD347 pKa = 3.22KK348 pKa = 10.95LGEE351 pKa = 4.04VHH353 pKa = 7.37ILQTDD358 pKa = 3.58VEE360 pKa = 4.59GRR362 pKa = 11.84AGISIRR368 pKa = 11.84RR369 pKa = 11.84LNGAGAPDD377 pKa = 3.55STIYY381 pKa = 10.37WDD383 pKa = 3.6DD384 pKa = 3.41TQMTGARR391 pKa = 11.84VNTTPVLSATDD402 pKa = 4.0GADD405 pKa = 3.39SAGGAHH411 pKa = 6.81GWAYY415 pKa = 10.98DD416 pKa = 3.61NNSWGNEE423 pKa = 3.62RR424 pKa = 11.84VIDD427 pKa = 3.77DD428 pKa = 3.89WTYY431 pKa = 11.11LPADD435 pKa = 4.63FGTGEE440 pKa = 3.84IQFGGRR446 pKa = 11.84CLSIDD451 pKa = 3.34KK452 pKa = 10.51TSDD455 pKa = 3.13FTEE458 pKa = 4.55DD459 pKa = 3.33SRR461 pKa = 11.84PGDD464 pKa = 3.45VVAYY468 pKa = 8.14TVTATNVGDD477 pKa = 3.61TDD479 pKa = 3.92YY480 pKa = 11.08TVADD484 pKa = 3.98PAVLFDD490 pKa = 5.02DD491 pKa = 4.98LSGVLDD497 pKa = 4.16DD498 pKa = 4.86ATYY501 pKa = 11.33NDD503 pKa = 4.17DD504 pKa = 3.99AAVVASDD511 pKa = 4.27GDD513 pKa = 4.3DD514 pKa = 3.6VPAPAFIEE522 pKa = 4.43PSHH525 pKa = 6.46LSWAGPLAAGEE536 pKa = 4.5SVEE539 pKa = 4.09LTYY542 pKa = 10.77TVTLGSAGDD551 pKa = 3.89GEE553 pKa = 4.8VANVAWQPVTTPPPGQVPPTPLCEE577 pKa = 4.15DD578 pKa = 3.57AVDD581 pKa = 4.0GRR583 pKa = 11.84DD584 pKa = 3.42PVTGEE589 pKa = 4.01ACDD592 pKa = 3.87GTVGEE597 pKa = 4.88LPRR600 pKa = 11.84LTIAKK605 pKa = 8.55TADD608 pKa = 3.25ATTLPTDD615 pKa = 3.88GDD617 pKa = 4.01EE618 pKa = 4.21VTYY621 pKa = 10.1TVTVTNEE628 pKa = 3.78GPGFYY633 pKa = 10.01TDD635 pKa = 4.74DD636 pKa = 4.21APATMTDD643 pKa = 3.19SLEE646 pKa = 4.14EE647 pKa = 4.05VLDD650 pKa = 4.12DD651 pKa = 4.97AEE653 pKa = 4.5FGEE656 pKa = 4.63ILAPAAGAEE665 pKa = 3.99FDD667 pKa = 4.29ADD669 pKa = 3.87AEE671 pKa = 4.28EE672 pKa = 4.68LTWSGPLAAGASIEE686 pKa = 3.73IRR688 pKa = 11.84YY689 pKa = 6.67TVVYY693 pKa = 10.14DD694 pKa = 3.77ATAGDD699 pKa = 4.03SVLTNTACVPEE710 pKa = 5.66ADD712 pKa = 4.04TLPGAEE718 pKa = 3.9SCATVRR724 pKa = 11.84IPGGRR729 pKa = 11.84LAVSKK734 pKa = 10.96SVDD737 pKa = 3.5PADD740 pKa = 3.89GSTVQSGQDD749 pKa = 3.14VTYY752 pKa = 9.73TLTFEE757 pKa = 4.46STGEE761 pKa = 4.15SEE763 pKa = 4.42VTVDD767 pKa = 5.61KK768 pKa = 11.48VDD770 pKa = 4.15DD771 pKa = 4.09LSGVLDD777 pKa = 3.87DD778 pKa = 5.14AEE780 pKa = 4.25FVEE783 pKa = 5.25GSIDD787 pKa = 3.68VSDD790 pKa = 4.14AAALTAVVDD799 pKa = 4.06GDD801 pKa = 3.78EE802 pKa = 4.21LAIDD806 pKa = 3.82GTIPAGQTVTVSYY819 pKa = 9.38TVTVAPYY826 pKa = 10.39AEE828 pKa = 4.4QGDD831 pKa = 4.23HH832 pKa = 6.22VLANVVQNPDD842 pKa = 3.53GTCGPEE848 pKa = 4.12GCPGTEE854 pKa = 3.36NPIRR858 pKa = 11.84HH859 pKa = 5.68FSITKK864 pKa = 7.76TAEE867 pKa = 3.83PVDD870 pKa = 4.45DD871 pKa = 3.96VTSGDD876 pKa = 3.56VVTYY880 pKa = 9.17TVTVTNDD887 pKa = 2.63GAADD891 pKa = 3.8YY892 pKa = 7.59TTDD895 pKa = 3.43APASVTDD902 pKa = 4.55DD903 pKa = 3.77LTDD906 pKa = 3.52VLDD909 pKa = 4.25DD910 pKa = 3.77AVYY913 pKa = 10.89DD914 pKa = 4.12GGATAVSSDD923 pKa = 3.38GSAVPAPQYY932 pKa = 10.91GEE934 pKa = 4.73PVLTWSGPVAAGEE947 pKa = 4.45SVEE950 pKa = 3.84ITYY953 pKa = 9.13TVTVTNLGDD962 pKa = 3.76HH963 pKa = 7.07DD964 pKa = 4.97LVNVAAPVCAPGEE977 pKa = 4.36DD978 pKa = 4.17CDD980 pKa = 4.36PPVSVEE986 pKa = 3.79ILLPSIVPAKK996 pKa = 10.63SSDD999 pKa = 3.71PASGEE1004 pKa = 4.03SLLAGDD1010 pKa = 4.12VVDD1013 pKa = 5.27YY1014 pKa = 9.92TLSWTNDD1021 pKa = 2.91GQAAGALDD1029 pKa = 3.89STDD1032 pKa = 4.36DD1033 pKa = 3.71LTGVLDD1039 pKa = 4.79DD1040 pKa = 5.5ADD1042 pKa = 3.73ITTAPQTSADD1052 pKa = 4.0GIEE1055 pKa = 4.67LVFDD1059 pKa = 4.75PDD1061 pKa = 3.44AGTLRR1066 pKa = 11.84VTGEE1070 pKa = 4.08LAPGEE1075 pKa = 4.67TVTVTYY1081 pKa = 10.6SVTVRR1086 pKa = 11.84PDD1088 pKa = 3.04GEE1090 pKa = 4.21RR1091 pKa = 11.84GDD1093 pKa = 4.71NIVGNVLVPDD1103 pKa = 4.05VPPFEE1108 pKa = 5.41CADD1111 pKa = 4.29ADD1113 pKa = 4.15PEE1115 pKa = 4.44CDD1117 pKa = 3.46PFVPPATSHH1126 pKa = 6.65PVGEE1130 pKa = 4.21LDD1132 pKa = 3.9DD1133 pKa = 4.29WKK1135 pKa = 9.8TVDD1138 pKa = 3.72PASGATVRR1146 pKa = 11.84AGQEE1150 pKa = 3.65VTYY1153 pKa = 9.26TLHH1156 pKa = 6.41FANIGEE1162 pKa = 4.23ADD1164 pKa = 3.33VDD1166 pKa = 4.03VARR1169 pKa = 11.84EE1170 pKa = 3.94DD1171 pKa = 3.78VLTQVLDD1178 pKa = 3.87DD1179 pKa = 4.14ATVVSPPEE1187 pKa = 4.1SSDD1190 pKa = 3.2AALSVFDD1197 pKa = 3.9VADD1200 pKa = 3.54GRR1202 pKa = 11.84FLVAGTLAPGQEE1214 pKa = 3.93ATVVYY1219 pKa = 8.73TVVVNDD1225 pKa = 3.87DD1226 pKa = 3.83GEE1228 pKa = 5.37RR1229 pKa = 11.84GDD1231 pKa = 5.14DD1232 pKa = 3.21RR1233 pKa = 11.84LGNFLVDD1240 pKa = 3.85EE1241 pKa = 4.79GEE1243 pKa = 4.34EE1244 pKa = 4.16PPATCEE1250 pKa = 3.76PTDD1253 pKa = 4.57AEE1255 pKa = 4.76RR1256 pKa = 11.84ADD1258 pKa = 3.87CTVNPVSNVQVTKK1271 pKa = 10.77SADD1274 pKa = 3.6PASGTTIAYY1283 pKa = 7.49DD1284 pKa = 3.31QRR1286 pKa = 11.84VTYY1289 pKa = 9.77TLTFRR1294 pKa = 11.84NVGTNPAAAPVAVDD1308 pKa = 3.75YY1309 pKa = 10.41TDD1311 pKa = 4.01HH1312 pKa = 7.15LSGVLDD1318 pKa = 4.06DD1319 pKa = 5.33AVLSGAPAASDD1330 pKa = 3.67PSLTAAVQGQAIRR1343 pKa = 11.84ITGTVATGDD1352 pKa = 4.05VVTVTYY1358 pKa = 9.44TVAPKK1363 pKa = 10.42AYY1365 pKa = 9.55DD1366 pKa = 3.48EE1367 pKa = 4.64QGDD1370 pKa = 4.1HH1371 pKa = 6.33VLANVVAQTGEE1382 pKa = 4.03EE1383 pKa = 4.6PICVPDD1389 pKa = 3.72SQLCTTHH1396 pKa = 6.96EE1397 pKa = 4.28LTPPPPLAATGGEE1410 pKa = 4.37VAASLLAAMMALLIGGGIFLAVSRR1434 pKa = 11.84RR1435 pKa = 11.84RR1436 pKa = 11.84RR1437 pKa = 11.84SVDD1440 pKa = 3.18QIGG1443 pKa = 3.11
MM1 pKa = 7.06ATSTRR6 pKa = 11.84ALIGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84PAARR17 pKa = 11.84PGRR20 pKa = 11.84SGEE23 pKa = 3.78RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84AGVAAAIVAALAVSLLGVALPSPALAAGNTPPSNAYY62 pKa = 10.06SNQWLYY68 pKa = 11.42AYY70 pKa = 10.33VEE72 pKa = 4.18AGEE75 pKa = 4.53TVVGTGNIEE84 pKa = 4.0QVVAPDD90 pKa = 3.54GTTALGPGTYY100 pKa = 10.34GPAAADD106 pKa = 3.91GVWRR110 pKa = 11.84IHH112 pKa = 6.68LPSQGTSIGYY122 pKa = 8.39SWTVDD127 pKa = 2.64VHH129 pKa = 6.45AAGSGQVIPGRR140 pKa = 11.84VWSDD144 pKa = 2.77RR145 pKa = 11.84YY146 pKa = 11.11YY147 pKa = 9.23MQQASGTANTVDD159 pKa = 3.63LNFWIVNNTGYY170 pKa = 10.98VYY172 pKa = 10.69AVEE175 pKa = 4.49LDD177 pKa = 4.12DD178 pKa = 4.22YY179 pKa = 11.55NGVNSIFEE187 pKa = 4.48ANSVGWADD195 pKa = 4.02EE196 pKa = 4.26ADD198 pKa = 4.9CIPPYY203 pKa = 10.76ASYY206 pKa = 10.4EE207 pKa = 4.05PPGGVGTTLPLPDD220 pKa = 4.66CGEE223 pKa = 4.11TYY225 pKa = 10.43RR226 pKa = 11.84VFFEE230 pKa = 4.84EE231 pKa = 4.7PADD234 pKa = 4.33DD235 pKa = 4.56LPQVSSTNLGSVNVRR250 pKa = 11.84PDD252 pKa = 3.52LLGPADD258 pKa = 4.32LAVDD262 pKa = 4.23DD263 pKa = 5.33LAFAPAGGGSATGTFTYY280 pKa = 10.36SINDD284 pKa = 3.82RR285 pKa = 11.84FTGGYY290 pKa = 9.12VLQIDD295 pKa = 3.75VDD297 pKa = 4.06GDD299 pKa = 3.64GSYY302 pKa = 11.07DD303 pKa = 3.59GQRR306 pKa = 11.84DD307 pKa = 3.18RR308 pKa = 11.84SIRR311 pKa = 11.84LGADD315 pKa = 3.02GSGSYY320 pKa = 10.44SYY322 pKa = 11.59AFDD325 pKa = 4.11GLDD328 pKa = 3.41GLGDD332 pKa = 4.56PIEE335 pKa = 4.84DD336 pKa = 4.05CTRR339 pKa = 11.84MGARR343 pKa = 11.84VFFDD347 pKa = 3.22KK348 pKa = 10.95LGEE351 pKa = 4.04VHH353 pKa = 7.37ILQTDD358 pKa = 3.58VEE360 pKa = 4.59GRR362 pKa = 11.84AGISIRR368 pKa = 11.84RR369 pKa = 11.84LNGAGAPDD377 pKa = 3.55STIYY381 pKa = 10.37WDD383 pKa = 3.6DD384 pKa = 3.41TQMTGARR391 pKa = 11.84VNTTPVLSATDD402 pKa = 4.0GADD405 pKa = 3.39SAGGAHH411 pKa = 6.81GWAYY415 pKa = 10.98DD416 pKa = 3.61NNSWGNEE423 pKa = 3.62RR424 pKa = 11.84VIDD427 pKa = 3.77DD428 pKa = 3.89WTYY431 pKa = 11.11LPADD435 pKa = 4.63FGTGEE440 pKa = 3.84IQFGGRR446 pKa = 11.84CLSIDD451 pKa = 3.34KK452 pKa = 10.51TSDD455 pKa = 3.13FTEE458 pKa = 4.55DD459 pKa = 3.33SRR461 pKa = 11.84PGDD464 pKa = 3.45VVAYY468 pKa = 8.14TVTATNVGDD477 pKa = 3.61TDD479 pKa = 3.92YY480 pKa = 11.08TVADD484 pKa = 3.98PAVLFDD490 pKa = 5.02DD491 pKa = 4.98LSGVLDD497 pKa = 4.16DD498 pKa = 4.86ATYY501 pKa = 11.33NDD503 pKa = 4.17DD504 pKa = 3.99AAVVASDD511 pKa = 4.27GDD513 pKa = 4.3DD514 pKa = 3.6VPAPAFIEE522 pKa = 4.43PSHH525 pKa = 6.46LSWAGPLAAGEE536 pKa = 4.5SVEE539 pKa = 4.09LTYY542 pKa = 10.77TVTLGSAGDD551 pKa = 3.89GEE553 pKa = 4.8VANVAWQPVTTPPPGQVPPTPLCEE577 pKa = 4.15DD578 pKa = 3.57AVDD581 pKa = 4.0GRR583 pKa = 11.84DD584 pKa = 3.42PVTGEE589 pKa = 4.01ACDD592 pKa = 3.87GTVGEE597 pKa = 4.88LPRR600 pKa = 11.84LTIAKK605 pKa = 8.55TADD608 pKa = 3.25ATTLPTDD615 pKa = 3.88GDD617 pKa = 4.01EE618 pKa = 4.21VTYY621 pKa = 10.1TVTVTNEE628 pKa = 3.78GPGFYY633 pKa = 10.01TDD635 pKa = 4.74DD636 pKa = 4.21APATMTDD643 pKa = 3.19SLEE646 pKa = 4.14EE647 pKa = 4.05VLDD650 pKa = 4.12DD651 pKa = 4.97AEE653 pKa = 4.5FGEE656 pKa = 4.63ILAPAAGAEE665 pKa = 3.99FDD667 pKa = 4.29ADD669 pKa = 3.87AEE671 pKa = 4.28EE672 pKa = 4.68LTWSGPLAAGASIEE686 pKa = 3.73IRR688 pKa = 11.84YY689 pKa = 6.67TVVYY693 pKa = 10.14DD694 pKa = 3.77ATAGDD699 pKa = 4.03SVLTNTACVPEE710 pKa = 5.66ADD712 pKa = 4.04TLPGAEE718 pKa = 3.9SCATVRR724 pKa = 11.84IPGGRR729 pKa = 11.84LAVSKK734 pKa = 10.96SVDD737 pKa = 3.5PADD740 pKa = 3.89GSTVQSGQDD749 pKa = 3.14VTYY752 pKa = 9.73TLTFEE757 pKa = 4.46STGEE761 pKa = 4.15SEE763 pKa = 4.42VTVDD767 pKa = 5.61KK768 pKa = 11.48VDD770 pKa = 4.15DD771 pKa = 4.09LSGVLDD777 pKa = 3.87DD778 pKa = 5.14AEE780 pKa = 4.25FVEE783 pKa = 5.25GSIDD787 pKa = 3.68VSDD790 pKa = 4.14AAALTAVVDD799 pKa = 4.06GDD801 pKa = 3.78EE802 pKa = 4.21LAIDD806 pKa = 3.82GTIPAGQTVTVSYY819 pKa = 9.38TVTVAPYY826 pKa = 10.39AEE828 pKa = 4.4QGDD831 pKa = 4.23HH832 pKa = 6.22VLANVVQNPDD842 pKa = 3.53GTCGPEE848 pKa = 4.12GCPGTEE854 pKa = 3.36NPIRR858 pKa = 11.84HH859 pKa = 5.68FSITKK864 pKa = 7.76TAEE867 pKa = 3.83PVDD870 pKa = 4.45DD871 pKa = 3.96VTSGDD876 pKa = 3.56VVTYY880 pKa = 9.17TVTVTNDD887 pKa = 2.63GAADD891 pKa = 3.8YY892 pKa = 7.59TTDD895 pKa = 3.43APASVTDD902 pKa = 4.55DD903 pKa = 3.77LTDD906 pKa = 3.52VLDD909 pKa = 4.25DD910 pKa = 3.77AVYY913 pKa = 10.89DD914 pKa = 4.12GGATAVSSDD923 pKa = 3.38GSAVPAPQYY932 pKa = 10.91GEE934 pKa = 4.73PVLTWSGPVAAGEE947 pKa = 4.45SVEE950 pKa = 3.84ITYY953 pKa = 9.13TVTVTNLGDD962 pKa = 3.76HH963 pKa = 7.07DD964 pKa = 4.97LVNVAAPVCAPGEE977 pKa = 4.36DD978 pKa = 4.17CDD980 pKa = 4.36PPVSVEE986 pKa = 3.79ILLPSIVPAKK996 pKa = 10.63SSDD999 pKa = 3.71PASGEE1004 pKa = 4.03SLLAGDD1010 pKa = 4.12VVDD1013 pKa = 5.27YY1014 pKa = 9.92TLSWTNDD1021 pKa = 2.91GQAAGALDD1029 pKa = 3.89STDD1032 pKa = 4.36DD1033 pKa = 3.71LTGVLDD1039 pKa = 4.79DD1040 pKa = 5.5ADD1042 pKa = 3.73ITTAPQTSADD1052 pKa = 4.0GIEE1055 pKa = 4.67LVFDD1059 pKa = 4.75PDD1061 pKa = 3.44AGTLRR1066 pKa = 11.84VTGEE1070 pKa = 4.08LAPGEE1075 pKa = 4.67TVTVTYY1081 pKa = 10.6SVTVRR1086 pKa = 11.84PDD1088 pKa = 3.04GEE1090 pKa = 4.21RR1091 pKa = 11.84GDD1093 pKa = 4.71NIVGNVLVPDD1103 pKa = 4.05VPPFEE1108 pKa = 5.41CADD1111 pKa = 4.29ADD1113 pKa = 4.15PEE1115 pKa = 4.44CDD1117 pKa = 3.46PFVPPATSHH1126 pKa = 6.65PVGEE1130 pKa = 4.21LDD1132 pKa = 3.9DD1133 pKa = 4.29WKK1135 pKa = 9.8TVDD1138 pKa = 3.72PASGATVRR1146 pKa = 11.84AGQEE1150 pKa = 3.65VTYY1153 pKa = 9.26TLHH1156 pKa = 6.41FANIGEE1162 pKa = 4.23ADD1164 pKa = 3.33VDD1166 pKa = 4.03VARR1169 pKa = 11.84EE1170 pKa = 3.94DD1171 pKa = 3.78VLTQVLDD1178 pKa = 3.87DD1179 pKa = 4.14ATVVSPPEE1187 pKa = 4.1SSDD1190 pKa = 3.2AALSVFDD1197 pKa = 3.9VADD1200 pKa = 3.54GRR1202 pKa = 11.84FLVAGTLAPGQEE1214 pKa = 3.93ATVVYY1219 pKa = 8.73TVVVNDD1225 pKa = 3.87DD1226 pKa = 3.83GEE1228 pKa = 5.37RR1229 pKa = 11.84GDD1231 pKa = 5.14DD1232 pKa = 3.21RR1233 pKa = 11.84LGNFLVDD1240 pKa = 3.85EE1241 pKa = 4.79GEE1243 pKa = 4.34EE1244 pKa = 4.16PPATCEE1250 pKa = 3.76PTDD1253 pKa = 4.57AEE1255 pKa = 4.76RR1256 pKa = 11.84ADD1258 pKa = 3.87CTVNPVSNVQVTKK1271 pKa = 10.77SADD1274 pKa = 3.6PASGTTIAYY1283 pKa = 7.49DD1284 pKa = 3.31QRR1286 pKa = 11.84VTYY1289 pKa = 9.77TLTFRR1294 pKa = 11.84NVGTNPAAAPVAVDD1308 pKa = 3.75YY1309 pKa = 10.41TDD1311 pKa = 4.01HH1312 pKa = 7.15LSGVLDD1318 pKa = 4.06DD1319 pKa = 5.33AVLSGAPAASDD1330 pKa = 3.67PSLTAAVQGQAIRR1343 pKa = 11.84ITGTVATGDD1352 pKa = 4.05VVTVTYY1358 pKa = 9.44TVAPKK1363 pKa = 10.42AYY1365 pKa = 9.55DD1366 pKa = 3.48EE1367 pKa = 4.64QGDD1370 pKa = 4.1HH1371 pKa = 6.33VLANVVAQTGEE1382 pKa = 4.03EE1383 pKa = 4.6PICVPDD1389 pKa = 3.72SQLCTTHH1396 pKa = 6.96EE1397 pKa = 4.28LTPPPPLAATGGEE1410 pKa = 4.37VAASLLAAMMALLIGGGIFLAVSRR1434 pKa = 11.84RR1435 pKa = 11.84RR1436 pKa = 11.84RR1437 pKa = 11.84SVDD1440 pKa = 3.18QIGG1443 pKa = 3.11
Molecular weight: 148.89 kDa
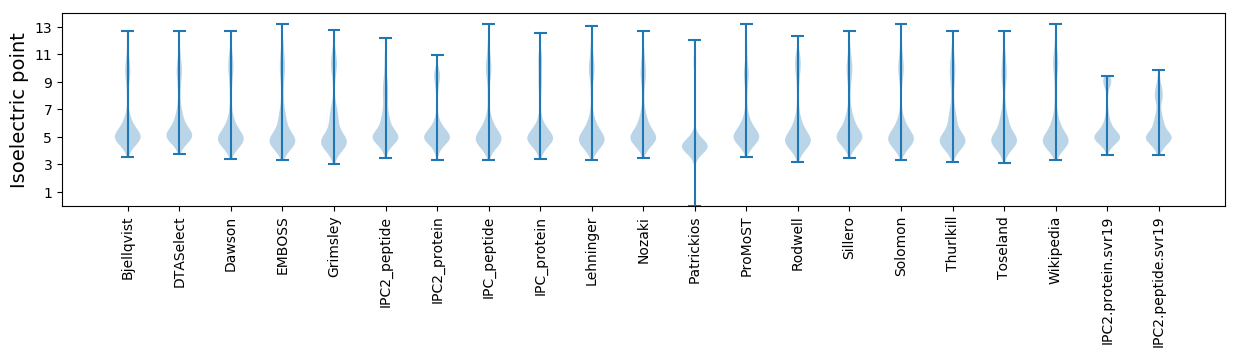
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4F7G5|A0A0U4F7G5_9MICO DNA helicase OS=Microbacterium sp. XT11 OX=367477 GN=AB663_001250 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.67GRR40 pKa = 11.84VEE42 pKa = 4.42LSAA45 pKa = 5.19
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.67GRR40 pKa = 11.84VEE42 pKa = 4.42LSAA45 pKa = 5.19
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
627981 |
38 |
2023 |
325.9 |
34.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.756 ± 0.08 | 0.447 ± 0.012 |
6.452 ± 0.048 | 5.814 ± 0.05 |
3.16 ± 0.033 | 8.877 ± 0.052 |
2.001 ± 0.026 | 4.525 ± 0.038 |
1.927 ± 0.035 | 9.999 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.851 ± 0.023 | 1.876 ± 0.03 |
5.372 ± 0.038 | 2.716 ± 0.026 |
7.396 ± 0.071 | 5.505 ± 0.046 |
6.012 ± 0.049 | 8.836 ± 0.055 |
1.504 ± 0.022 | 1.974 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
