
Wuhan spider virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.75
Get precalculated fractions of proteins
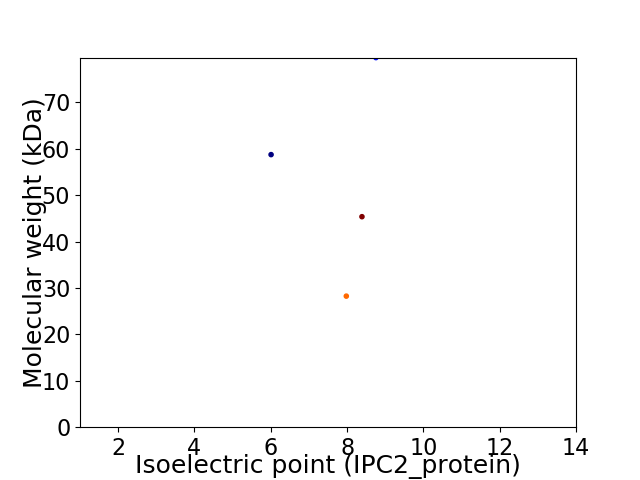
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGE8|A0A1L3KGE8_9VIRU RNA-directed RNA polymerase OS=Wuhan spider virus 8 OX=1923757 PE=4 SV=1
MM1 pKa = 6.88FVKK4 pKa = 9.79MAKK7 pKa = 9.8TIGRR11 pKa = 11.84QPKK14 pKa = 10.11ASWKK18 pKa = 10.0QMVASRR24 pKa = 11.84MSKK27 pKa = 9.34RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 9.69ALGAQYY38 pKa = 10.83YY39 pKa = 8.17WEE41 pKa = 4.2QGVTRR46 pKa = 11.84QDD48 pKa = 2.86AHH50 pKa = 7.36LSAFLKK56 pKa = 10.99LEE58 pKa = 4.33FYY60 pKa = 10.75KK61 pKa = 10.74EE62 pKa = 3.77EE63 pKa = 4.05KK64 pKa = 10.44LVEE67 pKa = 4.22KK68 pKa = 10.23EE69 pKa = 3.97CRR71 pKa = 11.84TIQHH75 pKa = 5.98RR76 pKa = 11.84RR77 pKa = 11.84VAYY80 pKa = 10.33NYY82 pKa = 11.17ALLRR86 pKa = 11.84WLCNIEE92 pKa = 3.55HH93 pKa = 6.2SMYY96 pKa = 10.98SKK98 pKa = 10.69LRR100 pKa = 11.84NVDD103 pKa = 3.35DD104 pKa = 3.87SPRR107 pKa = 11.84ILKK110 pKa = 9.56SVPPPMRR117 pKa = 11.84AFMVWRR123 pKa = 11.84AWEE126 pKa = 4.19SLGPTVRR133 pKa = 11.84CYY135 pKa = 11.0CLDD138 pKa = 3.14HH139 pKa = 7.17SRR141 pKa = 11.84FDD143 pKa = 3.42AHH145 pKa = 7.11VVRR148 pKa = 11.84DD149 pKa = 4.1WLEE152 pKa = 3.96MEE154 pKa = 3.83HH155 pKa = 7.23DD156 pKa = 4.55FYY158 pKa = 12.01KK159 pKa = 9.94MCSEE163 pKa = 4.44SRR165 pKa = 11.84RR166 pKa = 11.84EE167 pKa = 3.95LAWLLRR173 pKa = 11.84MQLRR177 pKa = 11.84AHH179 pKa = 6.46GRR181 pKa = 11.84TAGGATYY188 pKa = 10.55VQDD191 pKa = 4.27GKK193 pKa = 10.99RR194 pKa = 11.84SSGDD198 pKa = 3.17VNTASGNSADD208 pKa = 4.07NDD210 pKa = 3.75AMLLAHH216 pKa = 6.58MPDD219 pKa = 3.59NVCEE223 pKa = 4.22IVAEE227 pKa = 4.31VCVAAGFPDD236 pKa = 3.97LASRR240 pKa = 11.84ASDD243 pKa = 3.35YY244 pKa = 11.14RR245 pKa = 11.84FAVALLLQLLEE256 pKa = 4.35VEE258 pKa = 4.7LTPKK262 pKa = 10.49VGEE265 pKa = 4.22EE266 pKa = 4.07PPDD269 pKa = 3.89PVEE272 pKa = 4.17GTPLRR277 pKa = 11.84VITYY281 pKa = 10.36CDD283 pKa = 3.62GDD285 pKa = 4.15DD286 pKa = 4.66GLVLSNFKK294 pKa = 10.87LDD296 pKa = 3.59FFFALFGFSTEE307 pKa = 3.83ITVATSLEE315 pKa = 4.11QIEE318 pKa = 4.79FCQSKK323 pKa = 10.96LIMTSGGPAFARR335 pKa = 11.84DD336 pKa = 3.49PAKK339 pKa = 10.29VRR341 pKa = 11.84NMVMHH346 pKa = 6.79CRR348 pKa = 11.84NLPYY352 pKa = 10.03HH353 pKa = 5.29QRR355 pKa = 11.84EE356 pKa = 4.43GVLKK360 pKa = 10.45ASAFTEE366 pKa = 4.63YY367 pKa = 10.16IAQQGVWHH375 pKa = 6.16NQMLAYY381 pKa = 9.4KK382 pKa = 9.57CYY384 pKa = 9.45KK385 pKa = 10.02ACGSASPIFLNKK397 pKa = 9.86DD398 pKa = 3.56LAWRR402 pKa = 11.84CAALEE407 pKa = 3.99PVEE410 pKa = 3.97ITEE413 pKa = 4.67PKK415 pKa = 8.89WDD417 pKa = 5.01PIAIASFEE425 pKa = 4.23RR426 pKa = 11.84AWQFTLEE433 pKa = 4.12EE434 pKa = 4.42ALFFDD439 pKa = 4.29PVLRR443 pKa = 11.84TATEE447 pKa = 4.28GVVKK451 pKa = 10.64KK452 pKa = 10.88SSTLDD457 pKa = 3.51DD458 pKa = 3.44GTEE461 pKa = 3.86QKK463 pKa = 10.98EE464 pKa = 4.71CFLPDD469 pKa = 3.77HH470 pKa = 6.76GWPDD474 pKa = 3.53LVDD477 pKa = 3.55PTVCWRR483 pKa = 11.84CWAEE487 pKa = 4.12QKK489 pKa = 10.68PEE491 pKa = 3.56AAAAEE496 pKa = 4.33EE497 pKa = 4.17TTRR500 pKa = 11.84CCRR503 pKa = 11.84HH504 pKa = 6.01SCDD507 pKa = 4.39SNCGGWGGDD516 pKa = 3.46GG517 pKa = 3.63
MM1 pKa = 6.88FVKK4 pKa = 9.79MAKK7 pKa = 9.8TIGRR11 pKa = 11.84QPKK14 pKa = 10.11ASWKK18 pKa = 10.0QMVASRR24 pKa = 11.84MSKK27 pKa = 9.34RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 9.69ALGAQYY38 pKa = 10.83YY39 pKa = 8.17WEE41 pKa = 4.2QGVTRR46 pKa = 11.84QDD48 pKa = 2.86AHH50 pKa = 7.36LSAFLKK56 pKa = 10.99LEE58 pKa = 4.33FYY60 pKa = 10.75KK61 pKa = 10.74EE62 pKa = 3.77EE63 pKa = 4.05KK64 pKa = 10.44LVEE67 pKa = 4.22KK68 pKa = 10.23EE69 pKa = 3.97CRR71 pKa = 11.84TIQHH75 pKa = 5.98RR76 pKa = 11.84RR77 pKa = 11.84VAYY80 pKa = 10.33NYY82 pKa = 11.17ALLRR86 pKa = 11.84WLCNIEE92 pKa = 3.55HH93 pKa = 6.2SMYY96 pKa = 10.98SKK98 pKa = 10.69LRR100 pKa = 11.84NVDD103 pKa = 3.35DD104 pKa = 3.87SPRR107 pKa = 11.84ILKK110 pKa = 9.56SVPPPMRR117 pKa = 11.84AFMVWRR123 pKa = 11.84AWEE126 pKa = 4.19SLGPTVRR133 pKa = 11.84CYY135 pKa = 11.0CLDD138 pKa = 3.14HH139 pKa = 7.17SRR141 pKa = 11.84FDD143 pKa = 3.42AHH145 pKa = 7.11VVRR148 pKa = 11.84DD149 pKa = 4.1WLEE152 pKa = 3.96MEE154 pKa = 3.83HH155 pKa = 7.23DD156 pKa = 4.55FYY158 pKa = 12.01KK159 pKa = 9.94MCSEE163 pKa = 4.44SRR165 pKa = 11.84RR166 pKa = 11.84EE167 pKa = 3.95LAWLLRR173 pKa = 11.84MQLRR177 pKa = 11.84AHH179 pKa = 6.46GRR181 pKa = 11.84TAGGATYY188 pKa = 10.55VQDD191 pKa = 4.27GKK193 pKa = 10.99RR194 pKa = 11.84SSGDD198 pKa = 3.17VNTASGNSADD208 pKa = 4.07NDD210 pKa = 3.75AMLLAHH216 pKa = 6.58MPDD219 pKa = 3.59NVCEE223 pKa = 4.22IVAEE227 pKa = 4.31VCVAAGFPDD236 pKa = 3.97LASRR240 pKa = 11.84ASDD243 pKa = 3.35YY244 pKa = 11.14RR245 pKa = 11.84FAVALLLQLLEE256 pKa = 4.35VEE258 pKa = 4.7LTPKK262 pKa = 10.49VGEE265 pKa = 4.22EE266 pKa = 4.07PPDD269 pKa = 3.89PVEE272 pKa = 4.17GTPLRR277 pKa = 11.84VITYY281 pKa = 10.36CDD283 pKa = 3.62GDD285 pKa = 4.15DD286 pKa = 4.66GLVLSNFKK294 pKa = 10.87LDD296 pKa = 3.59FFFALFGFSTEE307 pKa = 3.83ITVATSLEE315 pKa = 4.11QIEE318 pKa = 4.79FCQSKK323 pKa = 10.96LIMTSGGPAFARR335 pKa = 11.84DD336 pKa = 3.49PAKK339 pKa = 10.29VRR341 pKa = 11.84NMVMHH346 pKa = 6.79CRR348 pKa = 11.84NLPYY352 pKa = 10.03HH353 pKa = 5.29QRR355 pKa = 11.84EE356 pKa = 4.43GVLKK360 pKa = 10.45ASAFTEE366 pKa = 4.63YY367 pKa = 10.16IAQQGVWHH375 pKa = 6.16NQMLAYY381 pKa = 9.4KK382 pKa = 9.57CYY384 pKa = 9.45KK385 pKa = 10.02ACGSASPIFLNKK397 pKa = 9.86DD398 pKa = 3.56LAWRR402 pKa = 11.84CAALEE407 pKa = 3.99PVEE410 pKa = 3.97ITEE413 pKa = 4.67PKK415 pKa = 8.89WDD417 pKa = 5.01PIAIASFEE425 pKa = 4.23RR426 pKa = 11.84AWQFTLEE433 pKa = 4.12EE434 pKa = 4.42ALFFDD439 pKa = 4.29PVLRR443 pKa = 11.84TATEE447 pKa = 4.28GVVKK451 pKa = 10.64KK452 pKa = 10.88SSTLDD457 pKa = 3.51DD458 pKa = 3.44GTEE461 pKa = 3.86QKK463 pKa = 10.98EE464 pKa = 4.71CFLPDD469 pKa = 3.77HH470 pKa = 6.76GWPDD474 pKa = 3.53LVDD477 pKa = 3.55PTVCWRR483 pKa = 11.84CWAEE487 pKa = 4.12QKK489 pKa = 10.68PEE491 pKa = 3.56AAAAEE496 pKa = 4.33EE497 pKa = 4.17TTRR500 pKa = 11.84CCRR503 pKa = 11.84HH504 pKa = 6.01SCDD507 pKa = 4.39SNCGGWGGDD516 pKa = 3.46GG517 pKa = 3.63
Molecular weight: 58.76 kDa
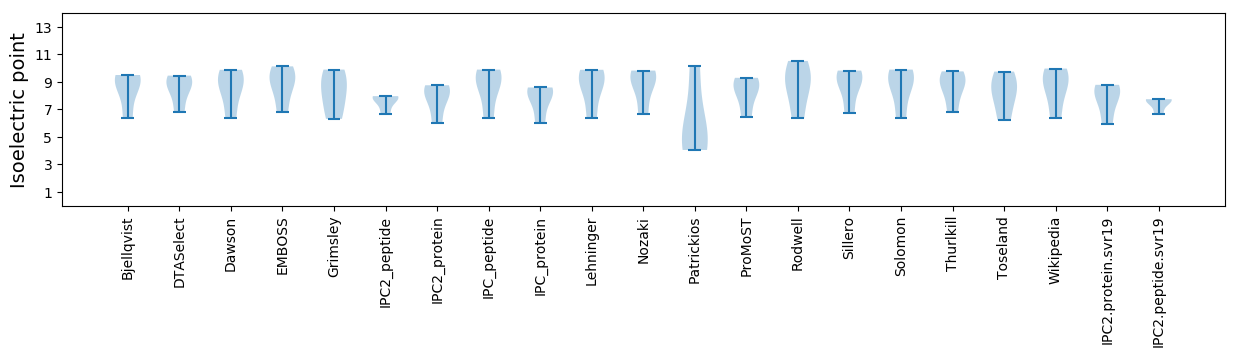
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGE8|A0A1L3KGE8_9VIRU RNA-directed RNA polymerase OS=Wuhan spider virus 8 OX=1923757 PE=4 SV=1
MM1 pKa = 7.41PAASFMEE8 pKa = 4.7RR9 pKa = 11.84IEE11 pKa = 4.12YY12 pKa = 10.54NITAAVTSAVLQSLGFVAPGTSSTPATSAAAAPPAPAKK50 pKa = 10.04VVKK53 pKa = 9.36TKK55 pKa = 9.12KK56 pKa = 8.43TPPKK60 pKa = 9.65AAPVKK65 pKa = 10.17AAAPPAPVKK74 pKa = 10.39AAAPPAPEE82 pKa = 5.04AEE84 pKa = 3.97VDD86 pKa = 3.64EE87 pKa = 4.96RR88 pKa = 11.84KK89 pKa = 8.14QTAKK93 pKa = 9.86QAKK96 pKa = 9.13KK97 pKa = 10.05AAKK100 pKa = 8.78QAKK103 pKa = 8.92KK104 pKa = 9.86AAKK107 pKa = 10.43GEE109 pKa = 4.27LLNKK113 pKa = 9.4AQQEE117 pKa = 4.38QEE119 pKa = 4.06KK120 pKa = 10.23SASSHH125 pKa = 4.29QEE127 pKa = 3.61KK128 pKa = 10.79PEE130 pKa = 4.02TEE132 pKa = 4.02ASAKK136 pKa = 8.82TRR138 pKa = 11.84LRR140 pKa = 11.84RR141 pKa = 11.84EE142 pKa = 3.63QRR144 pKa = 11.84KK145 pKa = 8.53KK146 pKa = 9.97QKK148 pKa = 10.11KK149 pKa = 7.92SKK151 pKa = 10.54KK152 pKa = 8.15PAKK155 pKa = 10.06QEE157 pKa = 3.7GFTPAKK163 pKa = 9.33PDD165 pKa = 3.31KK166 pKa = 10.39KK167 pKa = 11.17GKK169 pKa = 9.25GKK171 pKa = 10.49KK172 pKa = 9.4KK173 pKa = 9.5ATPEE177 pKa = 3.83TAGFVPLGEE186 pKa = 4.44SGAKK190 pKa = 9.48RR191 pKa = 11.84EE192 pKa = 4.21KK193 pKa = 10.95KK194 pKa = 9.84FGKK197 pKa = 10.33ADD199 pKa = 3.32ATHH202 pKa = 6.51YY203 pKa = 11.16KK204 pKa = 9.71MGWTKK209 pKa = 10.88EE210 pKa = 4.12EE211 pKa = 3.78QTKK214 pKa = 10.6ASSAFRR220 pKa = 11.84CDD222 pKa = 3.67LACVRR227 pKa = 11.84CNYY230 pKa = 8.59ATGNEE235 pKa = 3.81ICNNCRR241 pKa = 11.84KK242 pKa = 9.69FIKK245 pKa = 10.15LVCCSVEE252 pKa = 4.25KK253 pKa = 10.65PNEE256 pKa = 3.8AHH258 pKa = 6.55AKK260 pKa = 7.68WVAGGKK266 pKa = 9.92LGEE269 pKa = 4.76EE270 pKa = 4.45PPATYY275 pKa = 9.9IGAAEE280 pKa = 3.87RR281 pKa = 11.84RR282 pKa = 11.84VEE284 pKa = 4.04RR285 pKa = 11.84SRR287 pKa = 11.84GRR289 pKa = 11.84MRR291 pKa = 11.84VVCSGCDD298 pKa = 3.35TPIVDD303 pKa = 4.24WPRR306 pKa = 11.84FMLGWQHH313 pKa = 6.99AEE315 pKa = 3.92APSVRR320 pKa = 11.84IIMCKK325 pKa = 9.92RR326 pKa = 11.84KK327 pKa = 9.9ACRR330 pKa = 11.84EE331 pKa = 3.73PAAAGWLNPGSSFSPKK347 pKa = 9.23QVEE350 pKa = 4.41EE351 pKa = 4.96GPVIATADD359 pKa = 3.78EE360 pKa = 4.03LRR362 pKa = 11.84GISKK366 pKa = 10.42KK367 pKa = 10.51EE368 pKa = 3.44EE369 pKa = 4.48SKK371 pKa = 10.45PHH373 pKa = 5.11TRR375 pKa = 11.84GNPRR379 pKa = 11.84LQDD382 pKa = 3.69FLSEE386 pKa = 4.1NQFAALEE393 pKa = 4.16EE394 pKa = 4.2ASAAAGAKK402 pKa = 9.84GPRR405 pKa = 11.84VTPKK409 pKa = 9.84RR410 pKa = 11.84PKK412 pKa = 9.75KK413 pKa = 10.14ARR415 pKa = 11.84QSVEE419 pKa = 4.0AGSQTTEE426 pKa = 3.65PDD428 pKa = 2.95LRR430 pKa = 11.84NYY432 pKa = 10.13ISLDD436 pKa = 3.23AVRR439 pKa = 11.84TAIEE443 pKa = 4.13TGDD446 pKa = 3.77LEE448 pKa = 4.62RR449 pKa = 11.84LYY451 pKa = 11.32LEE453 pKa = 4.85AATHH457 pKa = 6.27AALSAAKK464 pKa = 10.16ASVDD468 pKa = 3.09AGTQTEE474 pKa = 4.81VPHH477 pKa = 5.93KK478 pKa = 10.19HH479 pKa = 6.21SVACGPDD486 pKa = 3.5AEE488 pKa = 4.53QSEE491 pKa = 4.92TPAAQDD497 pKa = 3.43SVASDD502 pKa = 3.63VAPSPSTSFAQAVRR516 pKa = 11.84GATASTSAAPQDD528 pKa = 3.64KK529 pKa = 10.8GVGQTPEE536 pKa = 3.88KK537 pKa = 9.91QQPKK541 pKa = 9.68ASKK544 pKa = 10.32SSKK547 pKa = 9.37PWPKK551 pKa = 10.64DD552 pKa = 3.1KK553 pKa = 10.75PVALAVGMSTLDD565 pKa = 3.22WVEE568 pKa = 5.35DD569 pKa = 4.72GIQEE573 pKa = 4.76LKK575 pKa = 11.13GKK577 pKa = 9.68FDD579 pKa = 3.93KK580 pKa = 11.07PHH582 pKa = 5.94GPKK585 pKa = 10.28ARR587 pKa = 11.84GVLLFSDD594 pKa = 4.3VPRR597 pKa = 11.84TYY599 pKa = 10.53SYY601 pKa = 11.48SGVKK605 pKa = 10.45LSPRR609 pKa = 11.84EE610 pKa = 3.86KK611 pKa = 10.27LPWMVSLEE619 pKa = 4.0RR620 pKa = 11.84LALKK624 pKa = 9.89YY625 pKa = 10.57LKK627 pKa = 10.54EE628 pKa = 4.08EE629 pKa = 4.35GGLDD633 pKa = 3.78VKK635 pKa = 10.29PTDD638 pKa = 3.61FGMLVVNDD646 pKa = 3.76YY647 pKa = 11.45GKK649 pKa = 10.35DD650 pKa = 3.32QGIPEE655 pKa = 4.5HH656 pKa = 7.0SDD658 pKa = 2.99NEE660 pKa = 3.98RR661 pKa = 11.84DD662 pKa = 3.28IDD664 pKa = 3.51QDD666 pKa = 3.57RR667 pKa = 11.84PIISLSVGGKK677 pKa = 9.73RR678 pKa = 11.84LMTFTDD684 pKa = 3.77LKK686 pKa = 10.78GEE688 pKa = 4.24VVEE691 pKa = 4.52EE692 pKa = 4.27VPLASWDD699 pKa = 3.94LVVMPPGSQKK709 pKa = 10.54LFHH712 pKa = 6.84HH713 pKa = 6.35KK714 pKa = 10.35VDD716 pKa = 3.92GEE718 pKa = 4.27GGPRR722 pKa = 11.84TNFTWRR728 pKa = 11.84AWKK731 pKa = 10.3SAGRR735 pKa = 11.84PHH737 pKa = 7.01PRR739 pKa = 3.24
MM1 pKa = 7.41PAASFMEE8 pKa = 4.7RR9 pKa = 11.84IEE11 pKa = 4.12YY12 pKa = 10.54NITAAVTSAVLQSLGFVAPGTSSTPATSAAAAPPAPAKK50 pKa = 10.04VVKK53 pKa = 9.36TKK55 pKa = 9.12KK56 pKa = 8.43TPPKK60 pKa = 9.65AAPVKK65 pKa = 10.17AAAPPAPVKK74 pKa = 10.39AAAPPAPEE82 pKa = 5.04AEE84 pKa = 3.97VDD86 pKa = 3.64EE87 pKa = 4.96RR88 pKa = 11.84KK89 pKa = 8.14QTAKK93 pKa = 9.86QAKK96 pKa = 9.13KK97 pKa = 10.05AAKK100 pKa = 8.78QAKK103 pKa = 8.92KK104 pKa = 9.86AAKK107 pKa = 10.43GEE109 pKa = 4.27LLNKK113 pKa = 9.4AQQEE117 pKa = 4.38QEE119 pKa = 4.06KK120 pKa = 10.23SASSHH125 pKa = 4.29QEE127 pKa = 3.61KK128 pKa = 10.79PEE130 pKa = 4.02TEE132 pKa = 4.02ASAKK136 pKa = 8.82TRR138 pKa = 11.84LRR140 pKa = 11.84RR141 pKa = 11.84EE142 pKa = 3.63QRR144 pKa = 11.84KK145 pKa = 8.53KK146 pKa = 9.97QKK148 pKa = 10.11KK149 pKa = 7.92SKK151 pKa = 10.54KK152 pKa = 8.15PAKK155 pKa = 10.06QEE157 pKa = 3.7GFTPAKK163 pKa = 9.33PDD165 pKa = 3.31KK166 pKa = 10.39KK167 pKa = 11.17GKK169 pKa = 9.25GKK171 pKa = 10.49KK172 pKa = 9.4KK173 pKa = 9.5ATPEE177 pKa = 3.83TAGFVPLGEE186 pKa = 4.44SGAKK190 pKa = 9.48RR191 pKa = 11.84EE192 pKa = 4.21KK193 pKa = 10.95KK194 pKa = 9.84FGKK197 pKa = 10.33ADD199 pKa = 3.32ATHH202 pKa = 6.51YY203 pKa = 11.16KK204 pKa = 9.71MGWTKK209 pKa = 10.88EE210 pKa = 4.12EE211 pKa = 3.78QTKK214 pKa = 10.6ASSAFRR220 pKa = 11.84CDD222 pKa = 3.67LACVRR227 pKa = 11.84CNYY230 pKa = 8.59ATGNEE235 pKa = 3.81ICNNCRR241 pKa = 11.84KK242 pKa = 9.69FIKK245 pKa = 10.15LVCCSVEE252 pKa = 4.25KK253 pKa = 10.65PNEE256 pKa = 3.8AHH258 pKa = 6.55AKK260 pKa = 7.68WVAGGKK266 pKa = 9.92LGEE269 pKa = 4.76EE270 pKa = 4.45PPATYY275 pKa = 9.9IGAAEE280 pKa = 3.87RR281 pKa = 11.84RR282 pKa = 11.84VEE284 pKa = 4.04RR285 pKa = 11.84SRR287 pKa = 11.84GRR289 pKa = 11.84MRR291 pKa = 11.84VVCSGCDD298 pKa = 3.35TPIVDD303 pKa = 4.24WPRR306 pKa = 11.84FMLGWQHH313 pKa = 6.99AEE315 pKa = 3.92APSVRR320 pKa = 11.84IIMCKK325 pKa = 9.92RR326 pKa = 11.84KK327 pKa = 9.9ACRR330 pKa = 11.84EE331 pKa = 3.73PAAAGWLNPGSSFSPKK347 pKa = 9.23QVEE350 pKa = 4.41EE351 pKa = 4.96GPVIATADD359 pKa = 3.78EE360 pKa = 4.03LRR362 pKa = 11.84GISKK366 pKa = 10.42KK367 pKa = 10.51EE368 pKa = 3.44EE369 pKa = 4.48SKK371 pKa = 10.45PHH373 pKa = 5.11TRR375 pKa = 11.84GNPRR379 pKa = 11.84LQDD382 pKa = 3.69FLSEE386 pKa = 4.1NQFAALEE393 pKa = 4.16EE394 pKa = 4.2ASAAAGAKK402 pKa = 9.84GPRR405 pKa = 11.84VTPKK409 pKa = 9.84RR410 pKa = 11.84PKK412 pKa = 9.75KK413 pKa = 10.14ARR415 pKa = 11.84QSVEE419 pKa = 4.0AGSQTTEE426 pKa = 3.65PDD428 pKa = 2.95LRR430 pKa = 11.84NYY432 pKa = 10.13ISLDD436 pKa = 3.23AVRR439 pKa = 11.84TAIEE443 pKa = 4.13TGDD446 pKa = 3.77LEE448 pKa = 4.62RR449 pKa = 11.84LYY451 pKa = 11.32LEE453 pKa = 4.85AATHH457 pKa = 6.27AALSAAKK464 pKa = 10.16ASVDD468 pKa = 3.09AGTQTEE474 pKa = 4.81VPHH477 pKa = 5.93KK478 pKa = 10.19HH479 pKa = 6.21SVACGPDD486 pKa = 3.5AEE488 pKa = 4.53QSEE491 pKa = 4.92TPAAQDD497 pKa = 3.43SVASDD502 pKa = 3.63VAPSPSTSFAQAVRR516 pKa = 11.84GATASTSAAPQDD528 pKa = 3.64KK529 pKa = 10.8GVGQTPEE536 pKa = 3.88KK537 pKa = 9.91QQPKK541 pKa = 9.68ASKK544 pKa = 10.32SSKK547 pKa = 9.37PWPKK551 pKa = 10.64DD552 pKa = 3.1KK553 pKa = 10.75PVALAVGMSTLDD565 pKa = 3.22WVEE568 pKa = 5.35DD569 pKa = 4.72GIQEE573 pKa = 4.76LKK575 pKa = 11.13GKK577 pKa = 9.68FDD579 pKa = 3.93KK580 pKa = 11.07PHH582 pKa = 5.94GPKK585 pKa = 10.28ARR587 pKa = 11.84GVLLFSDD594 pKa = 4.3VPRR597 pKa = 11.84TYY599 pKa = 10.53SYY601 pKa = 11.48SGVKK605 pKa = 10.45LSPRR609 pKa = 11.84EE610 pKa = 3.86KK611 pKa = 10.27LPWMVSLEE619 pKa = 4.0RR620 pKa = 11.84LALKK624 pKa = 9.89YY625 pKa = 10.57LKK627 pKa = 10.54EE628 pKa = 4.08EE629 pKa = 4.35GGLDD633 pKa = 3.78VKK635 pKa = 10.29PTDD638 pKa = 3.61FGMLVVNDD646 pKa = 3.76YY647 pKa = 11.45GKK649 pKa = 10.35DD650 pKa = 3.32QGIPEE655 pKa = 4.5HH656 pKa = 7.0SDD658 pKa = 2.99NEE660 pKa = 3.98RR661 pKa = 11.84DD662 pKa = 3.28IDD664 pKa = 3.51QDD666 pKa = 3.57RR667 pKa = 11.84PIISLSVGGKK677 pKa = 9.73RR678 pKa = 11.84LMTFTDD684 pKa = 3.77LKK686 pKa = 10.78GEE688 pKa = 4.24VVEE691 pKa = 4.52EE692 pKa = 4.27VPLASWDD699 pKa = 3.94LVVMPPGSQKK709 pKa = 10.54LFHH712 pKa = 6.84HH713 pKa = 6.35KK714 pKa = 10.35VDD716 pKa = 3.92GEE718 pKa = 4.27GGPRR722 pKa = 11.84TNFTWRR728 pKa = 11.84AWKK731 pKa = 10.3SAGRR735 pKa = 11.84PHH737 pKa = 7.01PRR739 pKa = 3.24
Molecular weight: 79.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1926 |
257 |
739 |
481.5 |
53.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.89 ± 0.778 | 2.077 ± 0.556 |
4.673 ± 0.404 | 6.646 ± 0.857 |
3.271 ± 0.511 | 6.802 ± 0.377 |
2.025 ± 0.163 | 2.7 ± 0.208 |
6.854 ± 1.775 | 7.788 ± 1.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.388 ± 0.399 | 2.752 ± 0.584 |
7.009 ± 0.797 | 3.946 ± 0.255 |
6.438 ± 0.421 | 6.75 ± 0.459 |
5.659 ± 0.663 | 5.919 ± 0.388 |
2.336 ± 0.387 | 2.077 ± 0.57 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
