
Blastococcus sp. CT_GayMR19
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
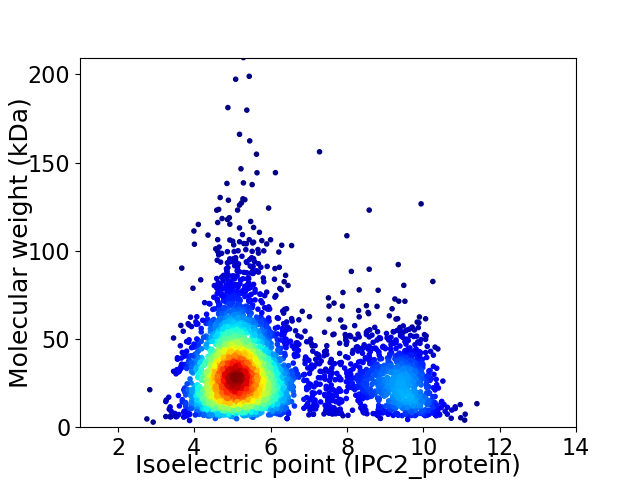
Virtual 2D-PAGE plot for 4236 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9PFB3|A0A4Y9PFB3_9ACTN S_2TMBeta domain-containing protein OS=Blastococcus sp. CT_GayMR19 OX=2559608 GN=E4P39_03420 PE=4 SV=1
MM1 pKa = 6.54TACGGSDD8 pKa = 3.4EE9 pKa = 4.61EE10 pKa = 5.01GSGEE14 pKa = 4.56GEE16 pKa = 3.97AAEE19 pKa = 4.06QAGGTFSMYY28 pKa = 10.18IGQPEE33 pKa = 4.15NPLVPGNTNEE43 pKa = 4.28TEE45 pKa = 4.22GGQVLDD51 pKa = 3.86SLFTGLVQYY60 pKa = 10.6DD61 pKa = 3.94AEE63 pKa = 4.33TNEE66 pKa = 4.16AVFSGVAASIEE77 pKa = 4.18SDD79 pKa = 4.73DD80 pKa = 3.86KK81 pKa = 10.86QTWTVTLNEE90 pKa = 3.94GWTFHH95 pKa = 7.57DD96 pKa = 4.41GTDD99 pKa = 3.23VTAQSFVDD107 pKa = 3.45AWNYY111 pKa = 7.22TAYY114 pKa = 10.76SPNAQGNSYY123 pKa = 10.41FFANVAGYY131 pKa = 11.23GDD133 pKa = 4.38LQAEE137 pKa = 4.27TDD139 pKa = 3.99DD140 pKa = 4.23NDD142 pKa = 3.83VVIAPPVAEE151 pKa = 4.12QMSGLRR157 pKa = 11.84VVDD160 pKa = 4.2DD161 pKa = 3.6LTFEE165 pKa = 4.43VEE167 pKa = 4.16LTSPYY172 pKa = 10.19AQWPTTVGYY181 pKa = 7.89TAFFPLPEE189 pKa = 4.67AFFADD194 pKa = 3.67PEE196 pKa = 4.76GFGEE200 pKa = 4.15QPIGNGPFQADD211 pKa = 3.82EE212 pKa = 4.49PFVPDD217 pKa = 4.22EE218 pKa = 5.07GITLTKK224 pKa = 10.11YY225 pKa = 10.42EE226 pKa = 4.74EE227 pKa = 4.33YY228 pKa = 11.0AGDD231 pKa = 3.75EE232 pKa = 4.24PANADD237 pKa = 3.4SVKK240 pKa = 9.7YY241 pKa = 9.92VVYY244 pKa = 10.73AEE246 pKa = 4.27QEE248 pKa = 3.98TAYY251 pKa = 9.84TDD253 pKa = 3.73LQGGQVDD260 pKa = 4.52IMDD263 pKa = 5.23TLPPDD268 pKa = 5.39AIASAPDD275 pKa = 3.47EE276 pKa = 4.6FGDD279 pKa = 4.63RR280 pKa = 11.84YY281 pKa = 10.53IEE283 pKa = 4.07TAQGDD288 pKa = 3.84ITSLGFPTYY297 pKa = 9.52DD298 pKa = 4.34QRR300 pKa = 11.84FADD303 pKa = 3.84PNVRR307 pKa = 11.84KK308 pKa = 9.59AFSMAIDD315 pKa = 3.7RR316 pKa = 11.84QAISDD321 pKa = 4.26AIFNGTRR328 pKa = 11.84TPATSFISPVVKK340 pKa = 10.04GHH342 pKa = 7.19RR343 pKa = 11.84DD344 pKa = 3.49DD345 pKa = 4.4ACEE348 pKa = 3.9VCEE351 pKa = 5.24LNVEE355 pKa = 4.36EE356 pKa = 5.29ANSLLDD362 pKa = 3.24EE363 pKa = 5.04AGFDD367 pKa = 3.37RR368 pKa = 11.84SQPVDD373 pKa = 2.84LWFNAGAGHH382 pKa = 6.42EE383 pKa = 4.27GWMEE387 pKa = 3.67AVGNQIRR394 pKa = 11.84EE395 pKa = 4.07NLDD398 pKa = 2.77VEE400 pKa = 4.44FQLRR404 pKa = 11.84GDD406 pKa = 4.18LQQSEE411 pKa = 4.59FLPLKK416 pKa = 9.59DD417 pKa = 3.82AKK419 pKa = 11.18GMTGPFRR426 pKa = 11.84DD427 pKa = 4.21AWVMDD432 pKa = 3.57YY433 pKa = 10.78PVAEE437 pKa = 4.34NFLGPLYY444 pKa = 10.73SSAALPPAGSNYY456 pKa = 9.03TFYY459 pKa = 11.44SNPEE463 pKa = 3.31FDD465 pKa = 3.93ALLVEE470 pKa = 5.29GNSASNDD477 pKa = 2.98EE478 pKa = 4.46DD479 pKa = 6.1AIAAYY484 pKa = 9.64QAAEE488 pKa = 4.93DD489 pKa = 3.98ILLEE493 pKa = 4.63DD494 pKa = 5.06FPSAPLFYY502 pKa = 10.61RR503 pKa = 11.84LNQGAHH509 pKa = 5.63SEE511 pKa = 4.1NVEE514 pKa = 4.0NVIIDD519 pKa = 3.32AFGRR523 pKa = 11.84IDD525 pKa = 3.23AAAVRR530 pKa = 11.84VVGG533 pKa = 4.26
MM1 pKa = 6.54TACGGSDD8 pKa = 3.4EE9 pKa = 4.61EE10 pKa = 5.01GSGEE14 pKa = 4.56GEE16 pKa = 3.97AAEE19 pKa = 4.06QAGGTFSMYY28 pKa = 10.18IGQPEE33 pKa = 4.15NPLVPGNTNEE43 pKa = 4.28TEE45 pKa = 4.22GGQVLDD51 pKa = 3.86SLFTGLVQYY60 pKa = 10.6DD61 pKa = 3.94AEE63 pKa = 4.33TNEE66 pKa = 4.16AVFSGVAASIEE77 pKa = 4.18SDD79 pKa = 4.73DD80 pKa = 3.86KK81 pKa = 10.86QTWTVTLNEE90 pKa = 3.94GWTFHH95 pKa = 7.57DD96 pKa = 4.41GTDD99 pKa = 3.23VTAQSFVDD107 pKa = 3.45AWNYY111 pKa = 7.22TAYY114 pKa = 10.76SPNAQGNSYY123 pKa = 10.41FFANVAGYY131 pKa = 11.23GDD133 pKa = 4.38LQAEE137 pKa = 4.27TDD139 pKa = 3.99DD140 pKa = 4.23NDD142 pKa = 3.83VVIAPPVAEE151 pKa = 4.12QMSGLRR157 pKa = 11.84VVDD160 pKa = 4.2DD161 pKa = 3.6LTFEE165 pKa = 4.43VEE167 pKa = 4.16LTSPYY172 pKa = 10.19AQWPTTVGYY181 pKa = 7.89TAFFPLPEE189 pKa = 4.67AFFADD194 pKa = 3.67PEE196 pKa = 4.76GFGEE200 pKa = 4.15QPIGNGPFQADD211 pKa = 3.82EE212 pKa = 4.49PFVPDD217 pKa = 4.22EE218 pKa = 5.07GITLTKK224 pKa = 10.11YY225 pKa = 10.42EE226 pKa = 4.74EE227 pKa = 4.33YY228 pKa = 11.0AGDD231 pKa = 3.75EE232 pKa = 4.24PANADD237 pKa = 3.4SVKK240 pKa = 9.7YY241 pKa = 9.92VVYY244 pKa = 10.73AEE246 pKa = 4.27QEE248 pKa = 3.98TAYY251 pKa = 9.84TDD253 pKa = 3.73LQGGQVDD260 pKa = 4.52IMDD263 pKa = 5.23TLPPDD268 pKa = 5.39AIASAPDD275 pKa = 3.47EE276 pKa = 4.6FGDD279 pKa = 4.63RR280 pKa = 11.84YY281 pKa = 10.53IEE283 pKa = 4.07TAQGDD288 pKa = 3.84ITSLGFPTYY297 pKa = 9.52DD298 pKa = 4.34QRR300 pKa = 11.84FADD303 pKa = 3.84PNVRR307 pKa = 11.84KK308 pKa = 9.59AFSMAIDD315 pKa = 3.7RR316 pKa = 11.84QAISDD321 pKa = 4.26AIFNGTRR328 pKa = 11.84TPATSFISPVVKK340 pKa = 10.04GHH342 pKa = 7.19RR343 pKa = 11.84DD344 pKa = 3.49DD345 pKa = 4.4ACEE348 pKa = 3.9VCEE351 pKa = 5.24LNVEE355 pKa = 4.36EE356 pKa = 5.29ANSLLDD362 pKa = 3.24EE363 pKa = 5.04AGFDD367 pKa = 3.37RR368 pKa = 11.84SQPVDD373 pKa = 2.84LWFNAGAGHH382 pKa = 6.42EE383 pKa = 4.27GWMEE387 pKa = 3.67AVGNQIRR394 pKa = 11.84EE395 pKa = 4.07NLDD398 pKa = 2.77VEE400 pKa = 4.44FQLRR404 pKa = 11.84GDD406 pKa = 4.18LQQSEE411 pKa = 4.59FLPLKK416 pKa = 9.59DD417 pKa = 3.82AKK419 pKa = 11.18GMTGPFRR426 pKa = 11.84DD427 pKa = 4.21AWVMDD432 pKa = 3.57YY433 pKa = 10.78PVAEE437 pKa = 4.34NFLGPLYY444 pKa = 10.73SSAALPPAGSNYY456 pKa = 9.03TFYY459 pKa = 11.44SNPEE463 pKa = 3.31FDD465 pKa = 3.93ALLVEE470 pKa = 5.29GNSASNDD477 pKa = 2.98EE478 pKa = 4.46DD479 pKa = 6.1AIAAYY484 pKa = 9.64QAAEE488 pKa = 4.93DD489 pKa = 3.98ILLEE493 pKa = 4.63DD494 pKa = 5.06FPSAPLFYY502 pKa = 10.61RR503 pKa = 11.84LNQGAHH509 pKa = 5.63SEE511 pKa = 4.1NVEE514 pKa = 4.0NVIIDD519 pKa = 3.32AFGRR523 pKa = 11.84IDD525 pKa = 3.23AAAVRR530 pKa = 11.84VVGG533 pKa = 4.26
Molecular weight: 57.66 kDa
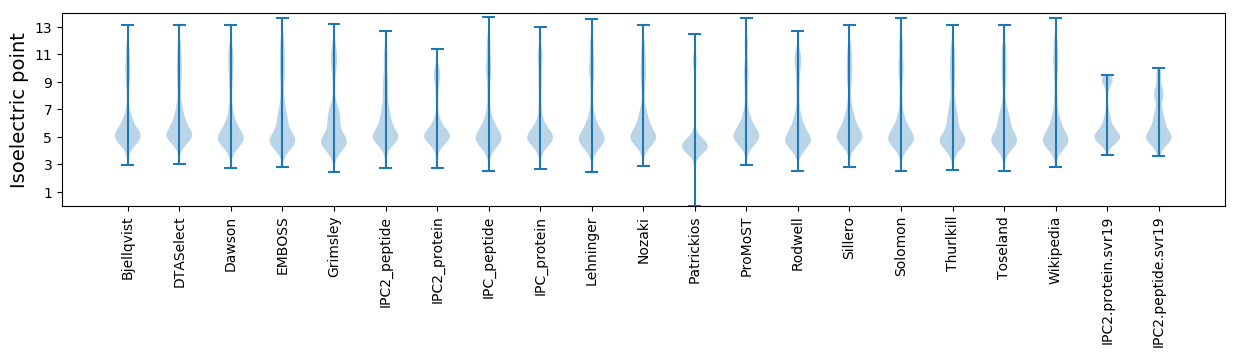
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9NRH4|A0A4Y9NRH4_9ACTN ATP-binding protein OS=Blastococcus sp. CT_GayMR19 OX=2559608 GN=E4P39_19845 PE=3 SV=1
MM1 pKa = 7.1PHH3 pKa = 6.5SGASATGRR11 pKa = 11.84WRR13 pKa = 11.84GPPQPLRR20 pKa = 11.84RR21 pKa = 11.84WPPPRR26 pKa = 11.84PPQPRR31 pKa = 11.84WPRR34 pKa = 11.84WPPRR38 pKa = 11.84LPAPPRR44 pKa = 11.84RR45 pKa = 11.84CRR47 pKa = 11.84RR48 pKa = 11.84TGRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VPLRR57 pKa = 11.84PHH59 pKa = 6.62SAPGPRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84PLRR73 pKa = 11.84PRR75 pKa = 11.84RR76 pKa = 11.84PWAPRR81 pKa = 11.84RR82 pKa = 11.84GRR84 pKa = 11.84RR85 pKa = 11.84PPPSWTAGSPPHH97 pKa = 6.8PAPAGLNTAPRR108 pKa = 11.84GRR110 pKa = 11.84AGRR113 pKa = 11.84PGAGG117 pKa = 3.17
MM1 pKa = 7.1PHH3 pKa = 6.5SGASATGRR11 pKa = 11.84WRR13 pKa = 11.84GPPQPLRR20 pKa = 11.84RR21 pKa = 11.84WPPPRR26 pKa = 11.84PPQPRR31 pKa = 11.84WPRR34 pKa = 11.84WPPRR38 pKa = 11.84LPAPPRR44 pKa = 11.84RR45 pKa = 11.84CRR47 pKa = 11.84RR48 pKa = 11.84TGRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VPLRR57 pKa = 11.84PHH59 pKa = 6.62SAPGPRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84PLRR73 pKa = 11.84PRR75 pKa = 11.84RR76 pKa = 11.84PWAPRR81 pKa = 11.84RR82 pKa = 11.84GRR84 pKa = 11.84RR85 pKa = 11.84PPPSWTAGSPPHH97 pKa = 6.8PAPAGLNTAPRR108 pKa = 11.84GRR110 pKa = 11.84AGRR113 pKa = 11.84PGAGG117 pKa = 3.17
Molecular weight: 13.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1347984 |
32 |
1957 |
318.2 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.048 ± 0.061 | 0.699 ± 0.008 |
6.196 ± 0.03 | 5.651 ± 0.032 |
2.663 ± 0.022 | 9.673 ± 0.037 |
1.99 ± 0.018 | 3.09 ± 0.028 |
1.514 ± 0.026 | 10.65 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.697 ± 0.016 | 1.552 ± 0.017 |
6.182 ± 0.032 | 2.651 ± 0.021 |
8.008 ± 0.042 | 5.054 ± 0.026 |
5.906 ± 0.028 | 9.564 ± 0.036 |
1.431 ± 0.017 | 1.781 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
